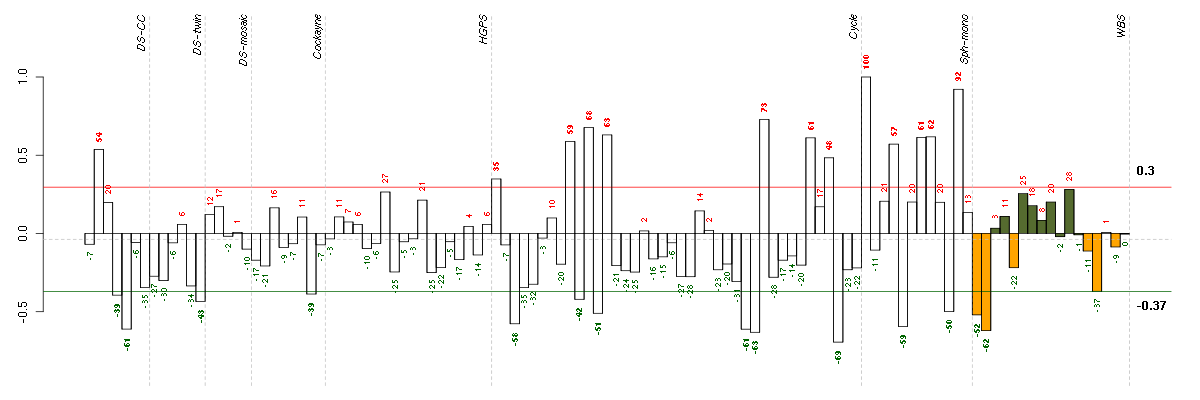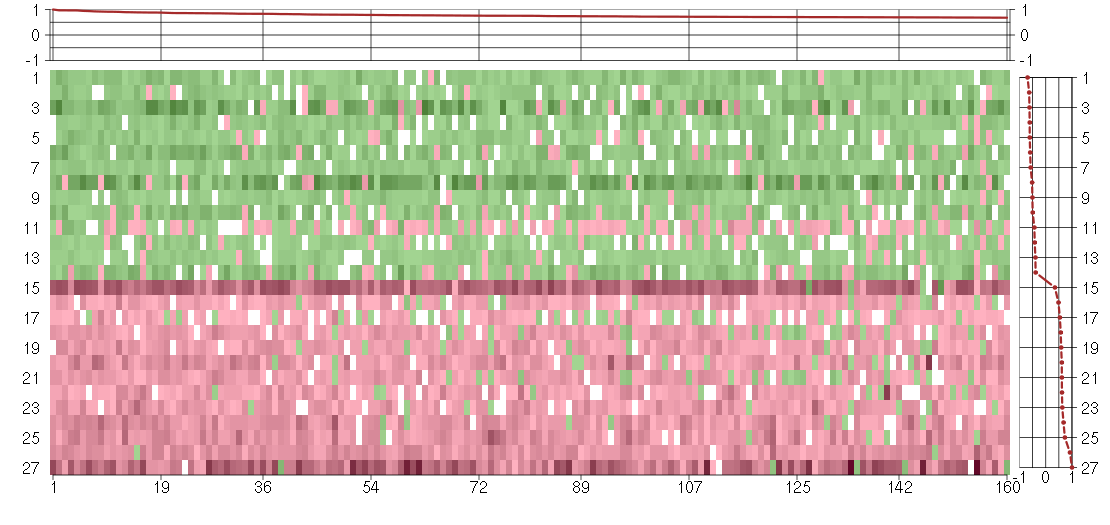

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.


| Id | Pvalue | ExpCount | Count | Size |
|---|---|---|---|---|
| miR-200bc/429 | 4.930e-02 | 13.77 | 31 | 576 |
| miR-17-5p/20/93.mr/106/519.d | 4.930e-02 | 15.27 | 33 | 639 |
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: 0.8 ACADSBacyl-Coenzyme A dehydrogenase, short/branched chain (205355_at), score: 0.7 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: 0.82 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: 0.89 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: 0.69 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.68 ANKHankylosis, progressive homolog (mouse) (220076_at), score: 0.88 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: 0.85 ANKRD12ankyrin repeat domain 12 (216550_x_at), score: 0.8 ANKRD46ankyrin repeat domain 46 (212731_at), score: 0.72 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.7 ARID4BAT rich interactive domain 4B (RBP1-like) (221230_s_at), score: 0.76 ATAD2BATPase family, AAA domain containing 2B (213387_at), score: 0.7 ATF1activating transcription factor 1 (222103_at), score: 0.68 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: 0.71 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (204194_at), score: 0.84 BAG5BCL2-associated athanogene 5 (202984_s_at), score: 0.68 BCORBCL6 co-repressor (219433_at), score: 0.73 BMP2bone morphogenetic protein 2 (205289_at), score: 0.8 BNIP2BCL2/adenovirus E1B 19kDa interacting protein 2 (209308_s_at), score: 0.86 BTAF1BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) (209430_at), score: 1 C10orf88chromosome 10 open reading frame 88 (219240_s_at), score: 0.83 C14orf138chromosome 14 open reading frame 138 (218940_at), score: 0.88 C1orf103chromosome 1 open reading frame 103 (220235_s_at), score: 0.74 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: 0.76 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: 0.74 CCDC47coiled-coil domain containing 47 (217814_at), score: 0.76 CCNJcyclin J (219470_x_at), score: 0.76 CCNT2cyclin T2 (204645_at), score: 0.74 CCT6Bchaperonin containing TCP1, subunit 6B (zeta 2) (206587_at), score: 0.75 CD302CD302 molecule (203799_at), score: 0.71 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: 0.7 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: 0.74 CHD1chromodomain helicase DNA binding protein 1 (204258_at), score: 0.75 CHSY1chondroitin sulfate synthase 1 (203044_at), score: 0.72 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: 0.69 CLCN3chloride channel 3 (201734_at), score: 0.72 COQ10Bcoenzyme Q10 homolog B (S. cerevisiae) (219397_at), score: 0.79 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.97 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.76 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: 0.71 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: 0.91 E2F3E2F transcription factor 3 (203693_s_at), score: 0.71 EDNRAendothelin receptor type A (204464_s_at), score: 0.75 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.69 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.71 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: 0.69 EREGepiregulin (205767_at), score: 0.73 EVI5ecotropic viral integration site 5 (209717_at), score: 0.89 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: 0.8 FLJ42627hypothetical LOC645644 (220352_x_at), score: 0.74 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: 0.79 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: 0.8 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: 0.68 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.88 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.73 GPR137BG protein-coupled receptor 137B (204137_at), score: 0.71 HECAheadcase homolog (Drosophila) (218603_at), score: 0.74 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.69 HK2hexokinase 2 (202934_at), score: 0.78 HS2ST1heparan sulfate 2-O-sulfotransferase 1 (203285_s_at), score: 0.77 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: 0.73 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.79 IL24interleukin 24 (206569_at), score: 0.69 INSRinsulin receptor (213792_s_at), score: 0.69 IREB2iron-responsive element binding protein 2 (214666_x_at), score: 0.7 IRGQimmunity-related GTPase family, Q (64488_at), score: 0.69 ITSN2intersectin 2 (209898_x_at), score: 0.77 JARID2jumonji, AT rich interactive domain 2 (203297_s_at), score: 0.82 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.92 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.86 KDSR3-ketodihydrosphingosine reductase (202419_at), score: 0.72 KIAA0247KIAA0247 (202181_at), score: 0.81 KIAA0562KIAA0562 (204075_s_at), score: 0.95 KIAA1024KIAA1024 (215081_at), score: 0.71 KIAA1128KIAA1128 (209379_s_at), score: 0.77 KLHL2kelch-like 2, Mayven (Drosophila) (219157_at), score: 0.69 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: 0.75 LEMD3LEM domain containing 3 (218604_at), score: 0.75 LOC644617hypothetical LOC644617 (221235_s_at), score: 0.77 LOC65998hypothetical protein LOC65998 (218641_at), score: 0.76 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: 0.71 LSM14ALSM14A, SCD6 homolog A (S. cerevisiae) (212131_at), score: 0.71 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: 0.68 MCM9minichromosome maintenance complex component 9 (219673_at), score: 0.72 MDN1MDN1, midasin homolog (yeast) (212693_at), score: 0.7 MED13Lmediator complex subunit 13-like (212207_at), score: 0.73 MEF2Amyocyte enhancer factor 2A (214684_at), score: 0.68 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.86 MORC3MORC family CW-type zinc finger 3 (213000_at), score: 0.82 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (220346_at), score: 0.71 MYNNmyoneurin (218926_at), score: 0.91 MYO6myosin VI (203216_s_at), score: 0.69 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: 0.77 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.78 NUMBnumb homolog (Drosophila) (207545_s_at), score: 0.8 NUPL1nucleoporin like 1 (204435_at), score: 0.88 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.82 PDCLphosducin-like (204449_at), score: 0.7 PELI1pellino homolog 1 (Drosophila) (218319_at), score: 0.83 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.78 PIGAphosphatidylinositol glycan anchor biosynthesis, class A (205281_s_at), score: 0.69 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: 0.73 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (202066_at), score: 0.68 PRDM2PR domain containing 2, with ZNF domain (203057_s_at), score: 0.69 PVRL3poliovirus receptor-related 3 (213325_at), score: 0.7 QTRTD1queuine tRNA-ribosyltransferase domain containing 1 (219178_at), score: 0.68 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: 0.74 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: 0.96 RB1CC1RB1-inducible coiled-coil 1 (202034_x_at), score: 0.85 RBM39RNA binding motif protein 39 (207941_s_at), score: 0.78 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: 0.7 RIPK2receptor-interacting serine-threonine kinase 2 (209544_at), score: 0.83 RNF11ring finger protein 11 (208924_at), score: 0.72 RNF38ring finger protein 38 (218528_s_at), score: 0.83 RUNX1runt-related transcription factor 1 (209360_s_at), score: 0.7 SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.71 SLC19A2solute carrier family 19 (thiamine transporter), member 2 (209681_at), score: 0.96 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: 0.75 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.85 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.83 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: 0.69 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.86 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: 0.71 SNRNP27small nuclear ribonucleoprotein 27kDa (U4/U6.U5) (212440_at), score: 0.72 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.69 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.76 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: 0.87 SSFA2sperm specific antigen 2 (202506_at), score: 0.75 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.9 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.77 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: 0.7 TBCCD1TBCC domain containing 1 (206451_at), score: 0.77 TDGthymine-DNA glycosylase (203743_s_at), score: 0.76 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: 0.78 TMEM66transmembrane protein 66 (200847_s_at), score: 0.73 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.71 TRAPPC10trafficking protein particle complex 10 (209412_at), score: 0.85 TRIM8tripartite motif-containing 8 (221012_s_at), score: 0.74 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: 0.84 UFM1ubiquitin-fold modifier 1 (218050_at), score: 0.97 USP12ubiquitin specific peptidase 12 (213327_s_at), score: 0.91 USP46ubiquitin specific peptidase 46 (203869_at), score: 0.72 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.7 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: 0.86 WDR44WD repeat domain 44 (219297_at), score: 0.7 WRNWerner syndrome (205667_at), score: 0.79 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.7 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.7 ZC3H11Azinc finger CCCH-type containing 11A (205788_s_at), score: 0.73 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: 0.83 ZCCHC2zinc finger, CCHC domain containing 2 (219062_s_at), score: 0.93 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.72 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.8 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: 0.7 ZMYM2zinc finger, MYM-type 2 (202778_s_at), score: 0.7 ZNF143zinc finger protein 143 (221873_at), score: 0.7 ZNF267zinc finger protein 267 (219540_at), score: 0.83 ZNF432zinc finger protein 432 (219848_s_at), score: 0.79 ZNF468zinc finger protein 468 (214751_at), score: 0.94
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21b 08-03.CEL | 5 | 1 | DS-CC | hgu133a | Down | DS-CC 5 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| 6Twin.CEL | 6 | 2 | DS-twin | hgu133plus2 | none | DS-twin 6 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| t21a 08-03.CEL | 4 | 1 | DS-CC | hgu133a | Down | DS-CC 4 |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515485651.cel | 1 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485851.cel | 11 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486231.cel | 30 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956083.cel | 2 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |