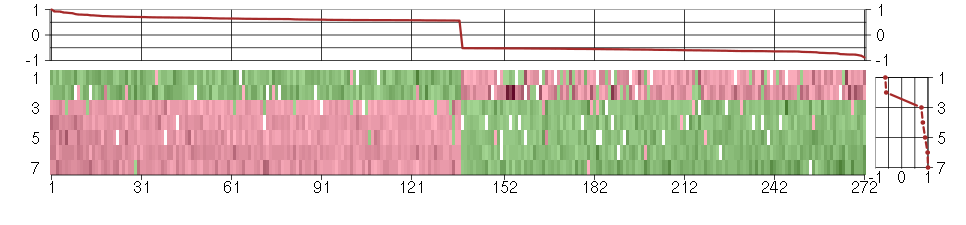

Under-expression is coded with green,
over-expression with red color.



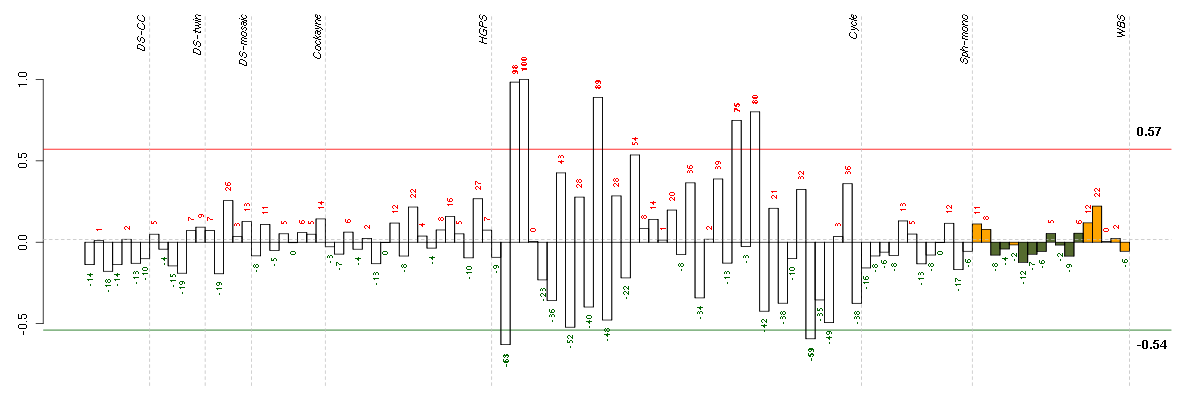
ABCA7ATP-binding cassette, sub-family A (ABC1), member 7 (219577_s_at), score: -0.76 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: -0.61 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: 0.71 ACTN3actinin, alpha 3 (206891_at), score: -0.55 ADAMTS12ADAM metallopeptidase with thrombospondin type 1 motif, 12 (221421_s_at), score: -0.52 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.57 ADORA1adenosine A1 receptor (216220_s_at), score: -0.56 AGTangiotensinogen (serpin peptidase inhibitor, clade A, member 8) (202834_at), score: -0.59 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.63 ARHGAP22Rho GTPase activating protein 22 (206298_at), score: 0.69 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.6 ARHGDIBRho GDP dissociation inhibitor (GDI) beta (201288_at), score: 0.64 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: -0.63 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: 0.58 ASPHD1aspartate beta-hydroxylase domain containing 1 (214993_at), score: -0.53 ATF5activating transcription factor 5 (204999_s_at), score: 0.66 ATMataxia telangiectasia mutated (210858_x_at), score: -0.55 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.74 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.6 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.7 BCL2L1BCL2-like 1 (215037_s_at), score: 0.65 BCL6B-cell CLL/lymphoma 6 (203140_at), score: -0.53 BTNL3butyrophilin-like 3 (217207_s_at), score: -0.52 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.52 C13orf34chromosome 13 open reading frame 34 (219544_at), score: 0.72 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.58 C16orf53chromosome 16 open reading frame 53 (218300_at), score: 0.58 C17orf91chromosome 17 open reading frame 91 (214696_at), score: -0.61 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.79 C1orf159chromosome 1 open reading frame 159 (219337_at), score: 0.59 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.61 C20orf27chromosome 20 open reading frame 27 (50314_i_at), score: 0.56 C2orf37chromosome 2 open reading frame 37 (220172_at), score: 0.71 C7orf44chromosome 7 open reading frame 44 (209445_x_at), score: 0.59 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.69 CAPN5calpain 5 (205166_at), score: 0.61 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.52 CCNFcyclin F (204826_at), score: 0.69 CD79BCD79b molecule, immunoglobulin-associated beta (205297_s_at), score: -0.63 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.65 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: 0.67 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.71 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.63 CDCA3cell division cycle associated 3 (221436_s_at), score: 0.58 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: 0.61 CENPEcentromere protein E, 312kDa (205046_at), score: 0.59 CHD1chromodomain helicase DNA binding protein 1 (204258_at), score: -0.52 CHRNB2cholinergic receptor, nicotinic, beta 2 (neuronal) (206635_at), score: -0.63 CHSY1chondroitin sulfate synthase 1 (203044_at), score: -0.52 CICcapicua homolog (Drosophila) (212784_at), score: 0.68 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.7 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.68 CNNM1cyclin M1 (220166_at), score: -0.53 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.73 COL16A1collagen, type XVI, alpha 1 (204345_at), score: -0.63 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.65 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: -0.58 CScitrate synthase (208660_at), score: 0.62 CTNScystinosis, nephropathic (36566_at), score: -0.55 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: -0.53 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: 0.71 DHCR77-dehydrocholesterol reductase (201790_s_at), score: -0.52 DHRS11dehydrogenase/reductase (SDR family) member 11 (218756_s_at), score: 0.58 DMBT1deleted in malignant brain tumors 1 (208250_s_at), score: -0.67 DMC1DMC1 dosage suppressor of mck1 homolog, meiosis-specific homologous recombination (yeast) (208382_s_at), score: 0.65 DNAJB2DnaJ (Hsp40) homolog, subfamily B, member 2 (202500_at), score: 0.59 DNAJC2DnaJ (Hsp40) homolog, subfamily C, member 2 (213097_s_at), score: -0.58 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.55 DUSP8dual specificity phosphatase 8 (206374_at), score: -0.56 ECM2extracellular matrix protein 2, female organ and adipocyte specific (206101_at), score: -0.55 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.62 ELK1ELK1, member of ETS oncogene family (210376_x_at), score: 0.65 EMCNendomucin (219436_s_at), score: 0.88 EPAGearly lymphoid activation protein (217050_at), score: -0.57 EPORerythropoietin receptor (215054_at), score: 0.64 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.6 EVLEnah/Vasp-like (217838_s_at), score: 0.63 EXOGendo/exonuclease (5'-3'), endonuclease G-like (205521_at), score: 0.63 F8A1coagulation factor VIII-associated (intronic transcript) 1 (203274_at), score: 0.59 FABP3fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) (214285_at), score: -0.63 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.68 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: -0.65 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: -0.52 FBP1fructose-1,6-bisphosphatase 1 (209696_at), score: -0.75 FER1L4fer-1-like 4 (C. elegans) (222245_s_at), score: -0.67 FGL1fibrinogen-like 1 (205305_at), score: -0.65 FKBP10FK506 binding protein 10, 65 kDa (219249_s_at), score: -0.54 FKRPfukutin related protein (219853_at), score: 0.57 FLJ21075hypothetical protein FLJ21075 (221172_at), score: -0.75 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: 0.77 FOSBFBJ murine osteosarcoma viral oncogene homolog B (202768_at), score: -0.52 FOXM1forkhead box M1 (202580_x_at), score: 0.57 FRG1FSHD region gene 1 (204145_at), score: -0.57 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.79 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.82 GADD45Bgrowth arrest and DNA-damage-inducible, beta (207574_s_at), score: -0.77 GBA3glucosidase, beta, acid 3 (cytosolic) (219954_s_at), score: -0.88 GDF9growth differentiation factor 9 (221314_at), score: -0.64 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.68 GNL3Lguanine nucleotide binding protein-like 3 (nucleolar)-like (205010_at), score: -0.62 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.6 GPR144G protein-coupled receptor 144 (216289_at), score: -0.58 GPR56G protein-coupled receptor 56 (212070_at), score: 0.69 GRB14growth factor receptor-bound protein 14 (206204_at), score: 0.66 H1F0H1 histone family, member 0 (208886_at), score: 0.64 H1FXH1 histone family, member X (204805_s_at), score: 0.65 HAB1B1 for mucin (215778_x_at), score: -0.71 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.61 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.57 HERC3hect domain and RLD 3 (206183_s_at), score: 0.69 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.72 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.6 HIVEP1human immunodeficiency virus type I enhancer binding protein 1 (204512_at), score: -0.53 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.88 HSD17B2hydroxysteroid (17-beta) dehydrogenase 2 (204818_at), score: -0.54 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.58 HUNKhormonally up-regulated Neu-associated kinase (219535_at), score: -0.53 IDI2isopentenyl-diphosphate delta isomerase 2 (217631_at), score: -0.64 IFI30interferon, gamma-inducible protein 30 (201422_at), score: 0.69 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (207901_at), score: 0.57 IL9Rinterleukin 9 receptor (214950_at), score: -0.64 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: 0.63 INTS1integrator complex subunit 1 (212212_s_at), score: 0.65 JAG2jagged 2 (32137_at), score: -0.58 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: 0.67 JUPjunction plakoglobin (201015_s_at), score: 0.76 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: -0.6 KIF15kinesin family member 15 (219306_at), score: 0.63 KIF26Bkinesin family member 26B (220002_at), score: -0.59 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: -0.77 LEPREL2leprecan-like 2 (204854_at), score: 0.66 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.63 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.56 LMF2lipase maturation factor 2 (212682_s_at), score: 0.59 LMO2LIM domain only 2 (rhombotin-like 1) (204249_s_at), score: 0.58 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: -0.52 LOC100133748similar to GTF2IRD2 protein (215569_at), score: -0.52 LOC149478hypothetical protein LOC149478 (215462_at), score: -0.59 LOC391132similar to hCG2041276 (216177_at), score: -0.56 LOC81691exonuclease NEF-sp (208107_s_at), score: 0.69 LPCAT3lysophosphatidylcholine acyltransferase 3 (202793_at), score: -0.62 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.59 LRP2low density lipoprotein-related protein 2 (205710_at), score: 0.63 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: -0.63 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.73 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.58 MEG3maternally expressed 3 (non-protein coding) (210794_s_at), score: -0.61 MEOX1mesenchyme homeobox 1 (205619_s_at), score: -0.65 MOBPmyelin-associated oligodendrocyte basic protein (210193_at), score: -0.6 MOCS1molybdenum cofactor synthesis 1 (211673_s_at), score: -0.56 MOGmyelin oligodendrocyte glycoprotein (214650_x_at), score: -0.55 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.64 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.77 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.79 MYH15myosin, heavy chain 15 (215331_at), score: -0.54 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.59 NCLNnicalin homolog (zebrafish) (222206_s_at), score: 0.62 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.92 NF1neurofibromin 1 (211094_s_at), score: 0.82 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: 0.58 NKRFNFKB repressing factor (205004_at), score: -0.54 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.56 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: -0.54 NR4A2nuclear receptor subfamily 4, group A, member 2 (216248_s_at), score: -0.53 NUAK1NUAK family, SNF1-like kinase, 1 (204589_at), score: -0.57 NUMA1nuclear mitotic apparatus protein 1 (214251_s_at), score: 0.57 NUPL2nucleoporin like 2 (204003_s_at), score: -0.55 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.7 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.58 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.58 OLFML2Bolfactomedin-like 2B (213125_at), score: 1 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.7 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.92 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: -0.52 PARVBparvin, beta (204629_at), score: 0.67 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.74 PHF7PHD finger protein 7 (215622_x_at), score: -0.64 PHLDA1pleckstrin homology-like domain, family A, member 1 (217997_at), score: 0.57 PHTF1putative homeodomain transcription factor 1 (215285_s_at), score: 0.67 PIM1pim-1 oncogene (209193_at), score: -0.52 PIP4K2Cphosphatidylinositol-5-phosphate 4-kinase, type II, gamma (218942_at), score: -0.52 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.59 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.58 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.63 PMLpromyelocytic leukemia (206503_x_at), score: 0.63 PNMAL1PNMA-like 1 (218824_at), score: 0.64 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: -0.64 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.6 PRRX1paired related homeobox 1 (205991_s_at), score: 0.64 PTCH1patched homolog 1 (Drosophila) (209815_at), score: -0.55 PTPN6protein tyrosine phosphatase, non-receptor type 6 (206687_s_at), score: -0.77 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.57 PXNpaxillin (211823_s_at), score: 0.86 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: -0.58 RASGRP1RAS guanyl releasing protein 1 (calcium and DAG-regulated) (205590_at), score: -0.64 RASGRP3RAS guanyl releasing protein 3 (calcium and DAG-regulated) (205801_s_at), score: 0.92 REG3Aregenerating islet-derived 3 alpha (205815_at), score: -0.71 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.57 RNF220ring finger protein 220 (219988_s_at), score: 0.57 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.54 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.73 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.58 RREB1ras responsive element binding protein 1 (203704_s_at), score: -0.57 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.59 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.6 SAA4serum amyloid A4, constitutive (207096_at), score: -0.6 SAP30LSAP30-like (219129_s_at), score: 0.58 SC4MOLsterol-C4-methyl oxidase-like (209146_at), score: -0.63 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.75 SCDstearoyl-CoA desaturase (delta-9-desaturase) (200832_s_at), score: -0.54 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.8 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: 0.57 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.53 SF1splicing factor 1 (208313_s_at), score: 0.6 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.58 SHROOM2shroom family member 2 (204967_at), score: -0.55 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.59 SLC11A2solute carrier family 11 (proton-coupled divalent metal ion transporters), member 2 (203124_s_at), score: -0.58 SLC26A10solute carrier family 26, member 10 (214951_at), score: -0.62 SLC29A1solute carrier family 29 (nucleoside transporters), member 1 (201801_s_at), score: 0.86 SLC2A1solute carrier family 2 (facilitated glucose transporter), member 1 (201250_s_at), score: -0.55 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: -0.66 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.57 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.62 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.68 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.57 SLC6A11solute carrier family 6 (neurotransmitter transporter, GABA), member 11 (207048_at), score: -0.72 SNTG1syntrophin, gamma 1 (220405_at), score: 0.63 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.6 SORBS1sorbin and SH3 domain containing 1 (218087_s_at), score: -0.61 SPATA2Lspermatogenesis associated 2-like (214965_at), score: -0.64 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.58 SRFserum response factor (c-fos serum response element-binding transcription factor) (202401_s_at), score: -0.53 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: -0.68 STIM1stromal interaction molecule 1 (202764_at), score: 0.59 STX3syntaxin 3 (209238_at), score: -0.53 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.53 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: -0.56 TDGthymine-DNA glycosylase (203743_s_at), score: -0.63 TGFBR2transforming growth factor, beta receptor II (70/80kDa) (207334_s_at), score: 0.8 TGM4transglutaminase 4 (prostate) (217566_s_at), score: -0.71 THRAthyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) (35846_at), score: -0.52 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.58 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.66 TMCO3transmembrane and coiled-coil domains 3 (220241_at), score: 0.61 TMEM121transmembrane protein 121 (219663_s_at), score: -0.53 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.59 TMEM48transmembrane protein 48 (218073_s_at), score: 0.57 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.55 TOM1L2target of myb1-like 2 (chicken) (214840_at), score: 0.65 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.52 TRAF5TNF receptor-associated factor 5 (204352_at), score: -0.7 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: -0.63 TRIM3tripartite motif-containing 3 (204911_s_at), score: 0.68 TTC33tetratricopeptide repeat domain 33 (219421_at), score: 0.57 TULP2tubby like protein 2 (206733_at), score: -0.63 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.61 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (211168_s_at), score: 0.59 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.52 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.74 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.59 WDR6WD repeat domain 6 (217734_s_at), score: 0.58 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: -0.62 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: 0.61 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.53 ZNF132zinc finger protein 132 (207402_at), score: 0.71 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: 0.63 ZNF20zinc finger protein 20 (213916_at), score: 0.59 ZNF281zinc finger protein 281 (218401_s_at), score: -0.67 ZNF510zinc finger protein 510 (206053_at), score: 0.7 ZNF652zinc finger protein 652 (205594_at), score: -0.71 ZNF783zinc finger family member 783 (221876_at), score: -0.56
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |