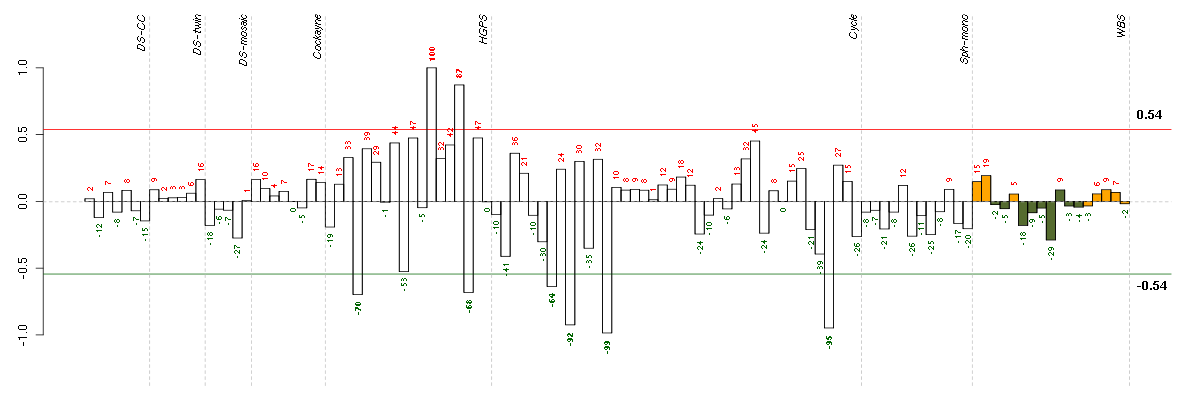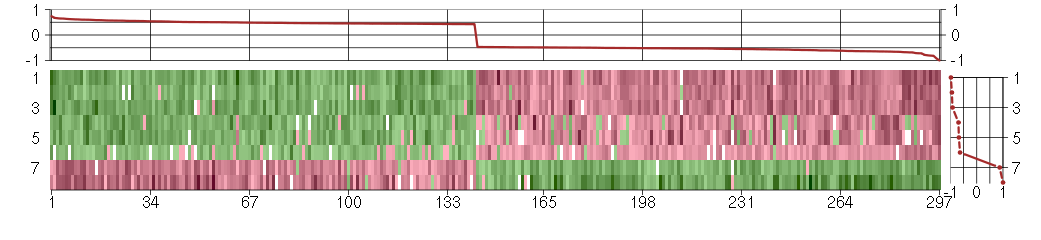

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
biological adhesion
The attachment of a cell or organism to a substrate or other organism.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
multicellular organismal metabolic process
The chemical reactions and pathways in multicellular organisms that occur at the tissue, organ, or organismal level. These processes, unlike cellular metabolism, can include transport of substances between cells when that transport is required.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
This term is the most general term possible
cell adhesion
The attachment of a cell, either to another cell or to an underlying substrate such as the extracellular matrix, via cell adhesion molecules.
multicellular organismal metabolic process
The chemical reactions and pathways in multicellular organisms that occur at the tissue, organ, or organismal level. These processes, unlike cellular metabolism, can include transport of substances between cells when that transport is required.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.43 ACANaggrecan (205679_x_at), score: -0.58 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.49 ADAMTS1ADAM metallopeptidase with thrombospondin type 1 motif, 1 (222162_s_at), score: -0.48 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: -0.51 ADD2adducin 2 (beta) (205268_s_at), score: 0.45 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.47 AHI1Abelson helper integration site 1 (221569_at), score: -0.62 AIM1absent in melanoma 1 (212543_at), score: 0.44 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (210962_s_at), score: -0.49 AKR1B1aldo-keto reductase family 1, member B1 (aldose reductase) (201272_at), score: 0.5 AKR1C1aldo-keto reductase family 1, member C1 (dihydrodiol dehydrogenase 1; 20-alpha (3-alpha)-hydroxysteroid dehydrogenase) (204151_x_at), score: 0.56 AKR1C2aldo-keto reductase family 1, member C2 (dihydrodiol dehydrogenase 2; bile acid binding protein; 3-alpha hydroxysteroid dehydrogenase, type III) (209699_x_at), score: 0.55 AMPHamphiphysin (205257_s_at), score: 0.49 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.44 ANO3anoctamin 3 (215241_at), score: -0.55 ANXA10annexin A10 (210143_at), score: -0.57 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.53 APOBEC3Bapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3B (206632_s_at), score: 0.46 ARL6IP1ADP-ribosylation factor-like 6 interacting protein 1 (211935_at), score: 0.53 ARSAarylsulfatase A (204443_at), score: 0.43 ARSJarylsulfatase family, member J (219973_at), score: -0.49 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (212361_s_at), score: -0.47 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.52 AUTS2autism susceptibility candidate 2 (212599_at), score: 0.5 AZI25-azacytidine induced 2 (218043_s_at), score: -0.47 B4GALT6UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 6 (206233_at), score: 0.45 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.46 BDKRB1bradykinin receptor B1 (207510_at), score: 0.43 BIN3bridging integrator 3 (222199_s_at), score: 0.61 BIRC5baculoviral IAP repeat-containing 5 (202095_s_at), score: 0.43 BMP6bone morphogenetic protein 6 (206176_at), score: -0.49 BSCL2Bernardinelli-Seip congenital lipodystrophy 2 (seipin) (208906_at), score: 0.46 BTG1B-cell translocation gene 1, anti-proliferative (200920_s_at), score: 0.45 C11orf51chromosome 11 open reading frame 51 (204218_at), score: 0.54 C11orf75chromosome 11 open reading frame 75 (219806_s_at), score: -0.57 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.64 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.47 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.64 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: -0.61 C2CD2C2 calcium-dependent domain containing 2 (212875_s_at), score: -0.49 C2orf34chromosome 2 open reading frame 34 (219617_at), score: -0.54 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.63 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.47 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.65 CAPN5calpain 5 (205166_at), score: 0.53 CARHSP1calcium regulated heat stable protein 1, 24kDa (218384_at), score: 0.44 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.51 CCDC46coiled-coil domain containing 46 (213644_at), score: 0.45 CCDC81coiled-coil domain containing 81 (220389_at), score: -0.5 CCDC85Bcoiled-coil domain containing 85B (204610_s_at), score: 0.43 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.48 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.59 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.59 CDK5RAP2CDK5 regulatory subunit associated protein 2 (220935_s_at), score: 0.57 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: 0.44 CDKN3cyclin-dependent kinase inhibitor 3 (209714_s_at), score: 0.48 CENPFcentromere protein F, 350/400ka (mitosin) (207828_s_at), score: 0.44 CGBchorionic gonadotropin, beta polypeptide (205387_s_at), score: 0.5 CHPT1choline phosphotransferase 1 (221675_s_at), score: 0.55 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.49 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.62 CKBcreatine kinase, brain (200884_at), score: 0.53 CLASP2cytoplasmic linker associated protein 2 (212306_at), score: -0.51 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.6 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.47 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.66 COBLL1COBL-like 1 (203642_s_at), score: -0.55 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.46 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.44 COL4A5collagen, type IV, alpha 5 (213110_s_at), score: -0.62 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.43 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (213379_at), score: 0.51 CORINcorin, serine peptidase (220356_at), score: -0.61 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.74 CTAGE5CTAGE family, member 5 (215930_s_at), score: -0.48 CUX1cut-like homeobox 1 (214743_at), score: -0.56 CYFIP2cytoplasmic FMR1 interacting protein 2 (215785_s_at), score: -0.51 DBNDD2dysbindin (dystrobrevin binding protein 1) domain containing 2 (218094_s_at), score: -0.57 DGCR6DiGeorge syndrome critical region gene 6 (208024_s_at), score: 0.5 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.55 DIDO1death inducer-obliterator 1 (218325_s_at), score: 0.51 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.55 DMDdystrophin (203881_s_at), score: -0.51 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.61 DNM1dynamin 1 (215116_s_at), score: 0.58 DOK5docking protein 5 (214844_s_at), score: -0.65 DPYSL3dihydropyrimidinase-like 3 (201430_s_at), score: 0.44 DSTdystonin (212253_x_at), score: 0.44 EAF2ELL associated factor 2 (219551_at), score: -0.67 EFHD1EF-hand domain family, member D1 (209343_at), score: -0.49 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.52 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.53 EMP2epithelial membrane protein 2 (204975_at), score: 0.6 EMX2empty spiracles homeobox 2 (221950_at), score: 0.48 ENPP1ectonucleotide pyrophosphatase/phosphodiesterase 1 (205066_s_at), score: 0.46 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.66 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.47 ERBB3v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (202454_s_at), score: -0.49 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.58 ESRRAestrogen-related receptor alpha (1487_at), score: -0.48 ETFBelectron-transfer-flavoprotein, beta polypeptide (202942_at), score: 0.52 EVI5ecotropic viral integration site 5 (209717_at), score: -0.57 F2RL2coagulation factor II (thrombin) receptor-like 2 (206795_at), score: -0.54 FADS2fatty acid desaturase 2 (202218_s_at), score: -0.49 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.65 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.58 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.53 FBXL7F-box and leucine-rich repeat protein 7 (213249_at), score: 0.53 FBXO31F-box protein 31 (219785_s_at), score: 0.51 FERMT2fermitin family homolog 2 (Drosophila) (214212_x_at), score: -0.49 FGF13fibroblast growth factor 13 (205110_s_at), score: 0.43 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.43 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.47 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.65 FOXM1forkhead box M1 (202580_x_at), score: 0.5 FTOfat mass and obesity associated (209702_at), score: 0.45 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.65 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.55 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: -0.66 GAS2L1growth arrest-specific 2 like 1 (31874_at), score: 0.51 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.46 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.55 GHRgrowth hormone receptor (205498_at), score: -0.68 GKglycerol kinase (207387_s_at), score: -0.47 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.55 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.68 GMPSguanine monphosphate synthetase (214431_at), score: 0.51 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (208813_at), score: 0.57 GPR116G protein-coupled receptor 116 (212950_at), score: -0.57 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.64 H1FXH1 histone family, member X (204805_s_at), score: 0.44 HDAC6histone deacetylase 6 (206846_s_at), score: 0.49 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.51 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.5 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.51 HOXC5homeobox C5 (206739_at), score: -0.53 HP1BP3heterochromatin protein 1, binding protein 3 (220633_s_at), score: 0.44 IL11RAinterleukin 11 receptor, alpha (204773_at), score: 0.43 IMAASLC7A5 pseudogene (208118_x_at), score: 0.46 INHBAinhibin, beta A (210511_s_at), score: -0.59 INSIG2insulin induced gene 2 (209566_at), score: -0.72 IRS1insulin receptor substrate 1 (204686_at), score: -0.51 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.46 IRX5iroquois homeobox 5 (210239_at), score: 0.54 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: 0.45 ITGA8integrin, alpha 8 (214265_at), score: -0.52 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.52 JAG1jagged 1 (Alagille syndrome) (216268_s_at), score: -0.49 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.71 KCNAB1potassium voltage-gated channel, shaker-related subfamily, beta member 1 (210078_s_at), score: -0.48 KIAA0355KIAA0355 (203288_at), score: -0.59 KIAA0562KIAA0562 (204075_s_at), score: -0.48 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.61 LGALS3BPlectin, galactoside-binding, soluble, 3 binding protein (200923_at), score: -0.48 LMNB2lamin B2 (216952_s_at), score: 0.45 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.48 LOC100133946similar to mature T-cell proliferations 1 (216862_s_at), score: 0.46 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.45 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.5 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.72 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.64 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.56 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.48 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.78 MGEA5meningioma expressed antigen 5 (hyaluronidase) (200898_s_at), score: 0.48 MGPmatrix Gla protein (202291_s_at), score: -0.52 MICBMHC class I polypeptide-related sequence B (206247_at), score: -0.49 MKI67antigen identified by monoclonal antibody Ki-67 (212022_s_at), score: 0.46 MLPHmelanophilin (218211_s_at), score: 0.47 MLXIPMLX interacting protein (202519_at), score: -0.53 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.56 MMP2matrix metallopeptidase 2 (gelatinase A, 72kDa gelatinase, 72kDa type IV collagenase) (201069_at), score: 0.56 MSX1msh homeobox 1 (205932_s_at), score: 0.56 MTCP1mature T-cell proliferation 1 (210212_x_at), score: 0.43 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.63 NCALDneurocalcin delta (211685_s_at), score: -0.48 NCAM1neural cell adhesion molecule 1 (212843_at), score: 0.51 NCAPD2non-SMC condensin I complex, subunit D2 (201774_s_at), score: 0.43 NDNnecdin homolog (mouse) (209550_at), score: 0.45 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.6 NEFLneurofilament, light polypeptide (221805_at), score: -0.52 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.5 NLGN4Yneuroligin 4, Y-linked (207703_at), score: -0.53 NOSIPnitric oxide synthase interacting protein (217950_at), score: 0.46 NPAS2neuronal PAS domain protein 2 (39549_at), score: -0.54 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.57 NPTX1neuronal pentraxin I (204684_at), score: -0.56 NRXN3neurexin 3 (205795_at), score: -0.92 NTF3neurotrophin 3 (206706_at), score: 0.52 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.47 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: 0.55 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.49 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.54 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.54 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.43 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.55 PCDH9protocadherin 9 (219737_s_at), score: -0.81 PDLIM3PDZ and LIM domain 3 (209621_s_at), score: -0.5 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.44 PHF7PHD finger protein 7 (215622_x_at), score: -0.55 PHLDA2pleckstrin homology-like domain, family A, member 2 (209803_s_at), score: -0.51 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.48 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.64 PLAC8placenta-specific 8 (219014_at), score: 0.48 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.52 PLATplasminogen activator, tissue (201860_s_at), score: -0.61 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.52 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.58 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (202066_at), score: -0.5 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.52 PRRX2paired related homeobox 2 (219729_at), score: 0.47 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: -0.63 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.66 PRSS3protease, serine, 3 (207463_x_at), score: -0.65 PTK2PTK2 protein tyrosine kinase 2 (208820_at), score: -0.64 PTPRZ1protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (204469_at), score: -0.53 PTTG1pituitary tumor-transforming 1 (203554_x_at), score: 0.43 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.44 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.5 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.53 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.5 RANBP17RAN binding protein 17 (219661_at), score: -0.55 RBM47RNA binding motif protein 47 (218035_s_at), score: 0.55 RELNreelin (205923_at), score: -0.63 RGS7regulator of G-protein signaling 7 (206290_s_at), score: -0.6 RHBDL2rhomboid, veinlet-like 2 (Drosophila) (219489_s_at), score: 0.6 RNF11ring finger protein 11 (208924_at), score: -0.56 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.53 RPS6KA5ribosomal protein S6 kinase, 90kDa, polypeptide 5 (204633_s_at), score: 0.57 RUNX3runt-related transcription factor 3 (204198_s_at), score: 0.51 SAP30LSAP30-like (219129_s_at), score: 0.48 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.46 SCN9Asodium channel, voltage-gated, type IX, alpha subunit (206950_at), score: -0.63 SDC1syndecan 1 (201287_s_at), score: 0.48 SEC24DSEC24 family, member D (S. cerevisiae) (202375_at), score: -0.53 SEPHS1selenophosphate synthetase 1 (208939_at), score: 0.45 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.63 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.59 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.5 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.5 SIP1survival of motor neuron protein interacting protein 1 (205063_at), score: -0.5 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.44 SLC1A1solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 (213664_at), score: -0.48 SLC24A3solute carrier family 24 (sodium/potassium/calcium exchanger), member 3 (57588_at), score: -0.61 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.68 SLC25A4solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 4 (202825_at), score: -0.54 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.56 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.51 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.57 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.59 SMURF2SMAD specific E3 ubiquitin protein ligase 2 (205596_s_at), score: -0.51 SNNstannin (218032_at), score: 0.67 SNTB1syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) (214708_at), score: -0.48 SOBPsine oculis binding protein homolog (Drosophila) (218974_at), score: -0.5 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.45 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.45 SRGAP2SLIT-ROBO Rho GTPase activating protein 2 (213329_at), score: 0.53 STK39serine threonine kinase 39 (STE20/SPS1 homolog, yeast) (202786_at), score: -0.52 STXBP2syntaxin binding protein 2 (209367_at), score: -0.52 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.8 TARDBPTAR DNA binding protein (200020_at), score: 0.43 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.52 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.58 TFPItissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) (209676_at), score: -0.64 THAP4THAP domain containing 4 (220417_s_at), score: 0.48 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: 0.44 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.47 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.5 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.62 TINAGL1tubulointerstitial nephritis antigen-like 1 (219058_x_at), score: -0.54 TMEM106Ctransmembrane protein 106C (201764_at), score: 0.61 TMEM186transmembrane protein 186 (204676_at), score: -0.48 TMEM2transmembrane protein 2 (218113_at), score: -0.52 TMEM45Atransmembrane protein 45A (219410_at), score: -0.5 TNS1tensin 1 (221748_s_at), score: -0.52 TNS3tensin 3 (217853_at), score: 0.46 TNXAtenascin XA pseudogene (213451_x_at), score: -0.51 TNXBtenascin XB (216333_x_at), score: -0.49 TPBGtrophoblast glycoprotein (203476_at), score: 0.45 TRIM14tripartite motif-containing 14 (203148_s_at), score: 0.49 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.48 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: -0.47 TSPAN12tetraspanin 12 (219274_at), score: -0.48 TTLL5tubulin tyrosine ligase-like family, member 5 (214672_at), score: 0.44 TWIST1twist homolog 1 (Drosophila) (213943_at), score: 0.46 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.81 WDR60WD repeat domain 60 (219251_s_at), score: -0.62 WHSC1Wolf-Hirschhorn syndrome candidate 1 (209053_s_at), score: 0.43 YTHDC2YTH domain containing 2 (213077_at), score: -0.51 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.45 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.47 ZCCHC14zinc finger, CCHC domain containing 14 (212655_at), score: 0.49 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.49 ZEB2zinc finger E-box binding homeobox 2 (203603_s_at), score: -0.56 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690231.cel | 4 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-GEOD-3860-raw-cel-1561690432.cel | 16 | 5 | HGPS | hgu133a | HGPS | AG10750 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690416.cel | 15 | 5 | HGPS | hgu133a | none | GM0316B |
| E-GEOD-3860-raw-cel-1561690360.cel | 12 | 5 | HGPS | hgu133a | none | GM0316B |