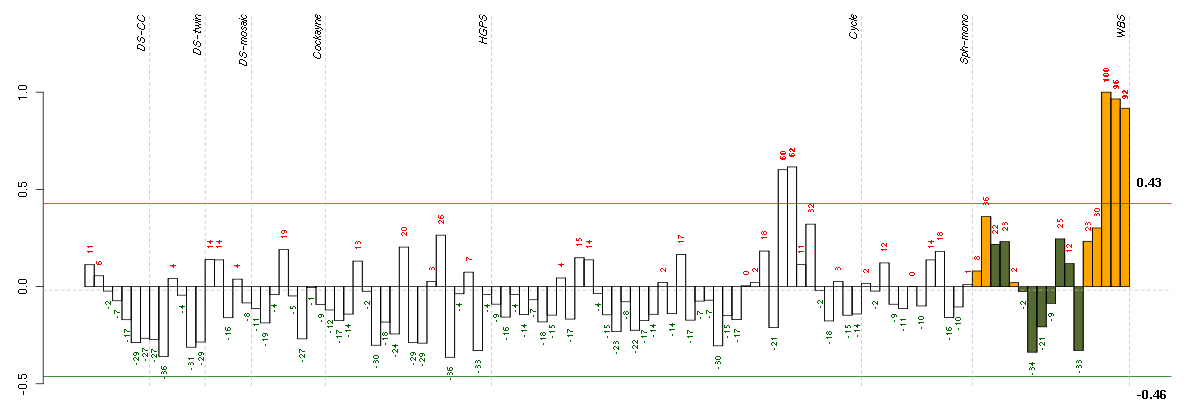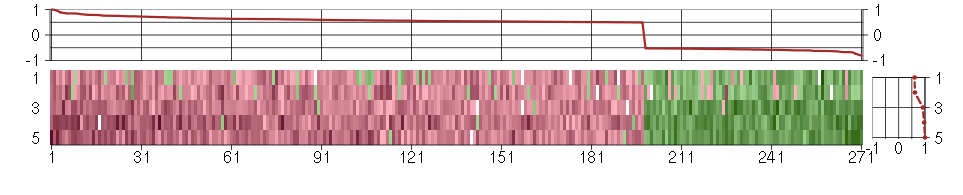

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
glutathione transferase activity
Catalysis of the reaction: R-X + glutathione = H-X + R-S-glutathione. R may be an aliphatic, aromatic or heterocyclic group; X may be a sulfate, nitrile or halide group.
transferase activity
Catalysis of the transfer of a group, e.g. a methyl group, glycosyl group, acyl group, phosphorus-containing, or other groups, from one compound (generally regarded as the donor) to another compound (generally regarded as the acceptor). Transferase is the systematic name for any enzyme of EC class 2.
transferase activity, transferring alkyl or aryl (other than methyl) groups
Catalysis of the transfer of an alkyl or aryl (but not methyl) group from one compound (donor) to another (acceptor).
all
This term is the most general term possible
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 00982 | 6.755e-03 | 1.07 | 7 | 36 | Drug metabolism - cytochrome P450 |
| 00980 | 7.864e-03 | 1.099 | 7 | 37 | Metabolism of xenobiotics by cytochrome P450 |
ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.64 ADCY9adenylate cyclase 9 (204497_at), score: 0.54 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.76 AGPAT51-acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) (218096_at), score: -0.62 AKAP6A kinase (PRKA) anchor protein 6 (205359_at), score: 0.74 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: -0.64 ALDH3B1aldehyde dehydrogenase 3 family, member B1 (211004_s_at), score: 0.62 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (221589_s_at), score: 0.63 AMD1adenosylmethionine decarboxylase 1 (201196_s_at), score: -0.56 ANGPT1angiopoietin 1 (205608_s_at), score: 0.58 ANKRD28ankyrin repeat domain 28 (213035_at), score: 0.56 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (209539_at), score: 0.65 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.66 ASPNasporin (219087_at), score: 0.72 B3GALT4UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 4 (210205_at), score: 0.6 B3GNT1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 1 (203188_at), score: 0.69 BAZ1Bbromodomain adjacent to zinc finger domain, 1B (208445_s_at), score: -0.57 BBS1Bardet-Biedl syndrome 1 (218471_s_at), score: 0.59 BCL7BB-cell CLL/lymphoma 7B (202518_at), score: -0.6 BDH23-hydroxybutyrate dehydrogenase, type 2 (218285_s_at), score: 0.54 BST1bone marrow stromal cell antigen 1 (205715_at), score: 0.52 BTDbiotinidase (204167_at), score: 0.51 BTG2BTG family, member 2 (201236_s_at), score: 0.56 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: 0.75 BTN3A2butyrophilin, subfamily 3, member A2 (209846_s_at), score: 0.71 BTN3A3butyrophilin, subfamily 3, member A3 (204820_s_at), score: 0.68 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.52 C10orf57chromosome 10 open reading frame 57 (218174_s_at), score: 0.63 C10orf72chromosome 10 open reading frame 72 (213381_at), score: 0.62 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.82 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.64 C17orf85chromosome 17 open reading frame 85 (218896_s_at), score: -0.58 C1orf115chromosome 1 open reading frame 115 (218546_at), score: -0.56 C1orf174chromosome 1 open reading frame 174 (222000_at), score: -0.57 C1orf50chromosome 1 open reading frame 50 (219406_at), score: 0.51 C2orf68chromosome 2 open reading frame 68 (221878_at), score: 0.55 C4orf18chromosome 4 open reading frame 18 (219872_at), score: 0.78 C4orf30chromosome 4 open reading frame 30 (219717_at), score: 0.61 C6orf35chromosome 6 open reading frame 35 (218453_s_at), score: 0.63 C7orf10chromosome 7 open reading frame 10 (219655_at), score: 0.54 C7orf58chromosome 7 open reading frame 58 (220032_at), score: 0.57 C9orf125chromosome 9 open reading frame 125 (213386_at), score: 0.6 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: 0.85 C9orf9chromosome 9 open reading frame 9 (220050_at), score: 0.5 CAMLGcalcium modulating ligand (203538_at), score: 0.53 CASP1caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) (211368_s_at), score: 0.53 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.51 CCNG1cyclin G1 (208796_s_at), score: 0.64 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: 0.7 CDH2cadherin 2, type 1, N-cadherin (neuronal) (203441_s_at), score: -0.52 CDH8cadherin 8, type 2 (217574_at), score: 0.68 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.51 CHST5carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 5 (219182_at), score: 0.59 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: 0.58 CLEC3BC-type lectin domain family 3, member B (205200_at), score: 0.57 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.53 COL8A2collagen, type VIII, alpha 2 (221900_at), score: 0.58 CPDcarboxypeptidase D (201942_s_at), score: 0.49 CRELD1cysteine-rich with EGF-like domains 1 (203368_at), score: 0.55 CRYBB2crystallin, beta B2 (206777_s_at), score: -0.55 CRYL1crystallin, lambda 1 (220753_s_at), score: 0.83 CTSCcathepsin C (201487_at), score: 0.5 CTSOcathepsin O (203758_at), score: 0.8 CXCR7chemokine (C-X-C motif) receptor 7 (212977_at), score: -0.53 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.62 DBC1deleted in bladder cancer 1 (205818_at), score: -0.73 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.53 DDX39DEAD (Asp-Glu-Ala-Asp) box polypeptide 39 (201584_s_at), score: -0.58 DHCR2424-dehydrocholesterol reductase (200862_at), score: 0.54 DHRS1dehydrogenase/reductase (SDR family) member 1 (213279_at), score: 0.55 DIAPH2diaphanous homolog 2 (Drosophila) (205603_s_at), score: 0.52 DLX2distal-less homeobox 2 (207147_at), score: -0.63 DMDdystrophin (203881_s_at), score: 0.51 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.58 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.65 DPYSL4dihydropyrimidinase-like 4 (205493_s_at), score: -0.63 DUSP12dual specificity phosphatase 12 (218576_s_at), score: -0.54 ECH1enoyl Coenzyme A hydratase 1, peroxisomal (200789_at), score: 0.52 EFHC2EF-hand domain (C-terminal) containing 2 (220591_s_at), score: 0.49 EFNA4ephrin-A4 (205107_s_at), score: -0.53 EGFL6EGF-like-domain, multiple 6 (219454_at), score: -0.55 EIF4A2eukaryotic translation initiation factor 4A, isoform 2 (200912_s_at), score: 0.58 EIF4Heukaryotic translation initiation factor 4H (206621_s_at), score: -0.6 EIF5eukaryotic translation initiation factor 5 (208290_s_at), score: -0.6 ENDOD1endonuclease domain containing 1 (212573_at), score: 0.55 ERAP1endoplasmic reticulum aminopeptidase 1 (214012_at), score: 0.56 F10coagulation factor X (205620_at), score: 0.7 FAM13Bfamily with sequence similarity 13, member B (218518_at), score: -0.54 FARSAphenylalanyl-tRNA synthetase, alpha subunit (216602_s_at), score: -0.56 FCHSD2FCH and double SH3 domains 2 (203620_s_at), score: 0.5 FDXRferredoxin reductase (207813_s_at), score: 0.52 FGD6FYVE, RhoGEF and PH domain containing 6 (219901_at), score: 0.53 FGFR1OPFGFR1 oncogene partner (205588_s_at), score: -0.59 FIG4FIG4 homolog (S. cerevisiae) (203656_at), score: 0.62 GAAglucosidase, alpha; acid (202812_at), score: 0.62 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: 0.65 GATA2GATA binding protein 2 (209710_at), score: -0.59 GBP2guanylate binding protein 2, interferon-inducible (202748_at), score: 0.68 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.99 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: 0.63 GLRBglycine receptor, beta (205280_at), score: 0.63 GPERG protein-coupled estrogen receptor 1 (210640_s_at), score: -0.53 GSNgelsolin (amyloidosis, Finnish type) (200696_s_at), score: 0.59 GSTM1glutathione S-transferase mu 1 (204550_x_at), score: 0.55 GSTM2glutathione S-transferase mu 2 (muscle) (204418_x_at), score: 0.54 GSTM3glutathione S-transferase mu 3 (brain) (202554_s_at), score: 0.5 GSTM4glutathione S-transferase mu 4 (204149_s_at), score: 0.51 GSTT2glutathione S-transferase theta 2 (205439_at), score: 0.56 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.85 HCFC1R1host cell factor C1 regulator 1 (XPO1 dependent) (45714_at), score: 0.53 HHLA3HERV-H LTR-associating 3 (220387_s_at), score: 0.73 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.66 HLXH2.0-like homeobox (214438_at), score: 0.57 HNRNPA2B1heterogeneous nuclear ribonucleoprotein A2/B1 (205292_s_at), score: -0.55 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (221480_at), score: -0.56 HOXA7homeobox A7 (206848_at), score: -0.63 HRASv-Ha-ras Harvey rat sarcoma viral oncogene homolog (212983_at), score: -0.56 HS1BP3HCLS1 binding protein 3 (219020_at), score: 0.52 HSPA5heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) (211936_at), score: -0.56 HSPB3heat shock 27kDa protein 3 (206375_s_at), score: 0.76 ID1inhibitor of DNA binding 1, dominant negative helix-loop-helix protein (208937_s_at), score: -0.81 ID3inhibitor of DNA binding 3, dominant negative helix-loop-helix protein (207826_s_at), score: -0.67 IFFO1intermediate filament family orphan 1 (36030_at), score: 0.55 IFI35interferon-induced protein 35 (209417_s_at), score: 0.69 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.56 IFITM1interferon induced transmembrane protein 1 (9-27) (214022_s_at), score: 0.59 IFITM2interferon induced transmembrane protein 2 (1-8D) (201315_x_at), score: 0.54 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.73 IGF1insulin-like growth factor 1 (somatomedin C) (209541_at), score: -0.66 IGFBP5insulin-like growth factor binding protein 5 (203424_s_at), score: 0.55 IL27RAinterleukin 27 receptor, alpha (205926_at), score: 0.73 IL6Rinterleukin 6 receptor (205945_at), score: 0.6 IMPAD1inositol monophosphatase domain containing 1 (218516_s_at), score: -0.53 ITIH5inter-alpha (globulin) inhibitor H5 (219064_at), score: -0.58 ITM2Bintegral membrane protein 2B (217732_s_at), score: 0.63 KCTD15potassium channel tetramerisation domain containing 15 (218553_s_at), score: -0.55 KIAA1107KIAA1107 (214098_at), score: 0.5 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.71 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: 0.75 LEPRleptin receptor (209894_at), score: 0.69 LEPREL2leprecan-like 2 (204854_at), score: 0.63 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.79 LHFPlipoma HMGIC fusion partner (218656_s_at), score: 0.75 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (218523_at), score: 0.61 LMF1lipase maturation factor 1 (219135_s_at), score: 0.65 LMOD1leiomodin 1 (smooth muscle) (203766_s_at), score: 0.72 LOC201229hypothetical protein LOC201229 (213195_at), score: 0.5 LOC645139hypothetical LOC645139 (209064_x_at), score: -0.55 LRRC32leucine rich repeat containing 32 (203835_at), score: 0.61 MACROD1MACRO domain containing 1 (219188_s_at), score: 0.58 MAGI2membrane associated guanylate kinase, WW and PDZ domain containing 2 (209737_at), score: 0.64 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (213178_s_at), score: 0.56 MEGF9multiple EGF-like-domains 9 (212830_at), score: 0.67 MEX3Dmex-3 homolog D (C. elegans) (91816_f_at), score: -0.58 MMP3matrix metallopeptidase 3 (stromelysin 1, progelatinase) (205828_at), score: -0.61 MPPE1metallophosphoesterase 1 (209858_x_at), score: 0.51 MR1major histocompatibility complex, class I-related (207565_s_at), score: 0.79 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: 0.54 N4BP2L1NEDD4 binding protein 2-like 1 (213375_s_at), score: 0.7 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (219217_at), score: 0.53 NAV2neuron navigator 2 (218330_s_at), score: -0.67 NCRNA00153non-protein coding RNA 153 (219961_s_at), score: 0.51 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.62 NEK9NIMA (never in mitosis gene a)- related kinase 9 (212299_at), score: 0.53 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: -0.53 NLGN1neuroligin 1 (205893_at), score: 0.53 NME3non-metastatic cells 3, protein expressed in (204862_s_at), score: 0.85 NOP56NOP56 ribonucleoprotein homolog (yeast) (200875_s_at), score: -0.53 NRCAMneuronal cell adhesion molecule (204105_s_at), score: -0.67 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: 0.52 OCEL1occludin/ELL domain containing 1 (205441_at), score: 0.65 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.75 PALLDpalladin, cytoskeletal associated protein (200906_s_at), score: 0.54 PAPOLGpoly(A) polymerase gamma (222273_at), score: 0.71 PAPPA2pappalysin 2 (213332_at), score: -0.68 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.5 PCYOX1Lprenylcysteine oxidase 1 like (218953_s_at), score: 0.79 PEX13peroxisomal biogenesis factor 13 (205246_at), score: -0.55 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.58 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: 0.59 PIGAphosphatidylinositol glycan anchor biosynthesis, class A (205281_s_at), score: -0.53 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: 0.5 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: 0.61 PLEKHO2pleckstrin homology domain containing, family O member 2 (204436_at), score: 0.54 PPANpeter pan homolog (Drosophila) (221649_s_at), score: -0.54 PPCDCphosphopantothenoylcysteine decarboxylase (219066_at), score: -0.58 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta isoform (221772_s_at), score: -0.52 PPP2R5Aprotein phosphatase 2, regulatory subunit B', alpha isoform (202187_s_at), score: 0.51 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: 0.85 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.62 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.75 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.57 PTPRAprotein tyrosine phosphatase, receptor type, A (213799_s_at), score: 0.49 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.51 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.54 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.61 R3HDM2R3H domain containing 2 (203831_at), score: 0.52 RAB22ARAB22A, member RAS oncogene family (218360_at), score: -0.58 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: -0.57 RCBTB2regulator of chromosome condensation (RCC1) and BTB (POZ) domain containing protein 2 (204759_at), score: 0.67 RFC2replication factor C (activator 1) 2, 40kDa (203696_s_at), score: -0.6 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.5 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.52 RHOQras homolog gene family, member Q (212119_at), score: 0.52 RNMTRNA (guanine-7-) methyltransferase (202684_s_at), score: -0.59 SAMD14sterile alpha motif domain containing 14 (213866_at), score: 0.57 SESN1sestrin 1 (218346_s_at), score: 0.57 SGSM2small G protein signaling modulator 2 (36129_at), score: 0.49 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.52 SLC20A1solute carrier family 20 (phosphate transporter), member 1 (201920_at), score: -0.64 SLC22A18solute carrier family 22, member 18 (204981_at), score: 0.5 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.62 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.53 SLC27A3solute carrier family 27 (fatty acid transporter), member 3 (222217_s_at), score: 0.64 SLC43A1solute carrier family 43, member 1 (204394_at), score: 0.64 SLC46A3solute carrier family 46, member 3 (214719_at), score: 1 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: 0.63 SLC4A2solute carrier family 4, anion exchanger, member 2 (erythrocyte membrane protein band 3-like 1) (202111_at), score: -0.53 SMAD7SMAD family member 7 (204790_at), score: -0.56 SMARCA2SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 (212257_s_at), score: 0.51 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.54 SOD3superoxide dismutase 3, extracellular (205236_x_at), score: 0.55 SORT1sortilin 1 (212807_s_at), score: 0.56 SPON1spondin 1, extracellular matrix protein (209436_at), score: -0.78 SREBF2sterol regulatory element binding transcription factor 2 (201247_at), score: 0.54 SSH3slingshot homolog 3 (Drosophila) (219241_x_at), score: 0.52 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.66 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.95 STAT4signal transducer and activator of transcription 4 (206118_at), score: 0.51 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.62 SYNPOsynaptopodin (202796_at), score: 0.56 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.58 TBC1D12TBC1 domain family, member 12 (221858_at), score: 0.73 TBC1D5TBC1 domain family, member 5 (201813_s_at), score: 0.54 TBL2transducin (beta)-like 2 (212685_s_at), score: -0.53 TBX1T-box 1 (211273_s_at), score: 0.54 TBX3T-box 3 (219682_s_at), score: -0.53 THEM2thioesterase superfamily member 2 (204565_at), score: 0.52 THNSL2threonine synthase-like 2 (S. cerevisiae) (219044_at), score: 0.51 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.53 TIMP3TIMP metallopeptidase inhibitor 3 (201149_s_at), score: -0.55 TM4SF20transmembrane 4 L six family member 20 (220639_at), score: 0.62 TMEM159transmembrane protein 159 (213272_s_at), score: 0.61 TMEM187transmembrane protein 187 (204340_at), score: 0.59 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.59 TNFSF12tumor necrosis factor (ligand) superfamily, member 12 (205611_at), score: 0.55 TOMM7translocase of outer mitochondrial membrane 7 homolog (yeast) (201812_s_at), score: 0.72 TP53TG1TP53 target 1 (non-protein coding) (210241_s_at), score: 0.51 TPCN1two pore segment channel 1 (217914_at), score: 0.57 TPRKBTP53RK binding protein (219030_at), score: -0.54 TRA2Btransformer 2 beta homolog (Drosophila) (200892_s_at), score: -0.63 TRAPPC6Atrafficking protein particle complex 6A (204985_s_at), score: 0.79 TRIM22tripartite motif-containing 22 (213293_s_at), score: 0.54 TSC22D1TSC22 domain family, member 1 (215111_s_at), score: 0.54 TUBA1Ctubulin, alpha 1c (211750_x_at), score: -0.54 TUBA3Ctubulin, alpha 3c (210527_x_at), score: -0.6 TUBA3Dtubulin, alpha 3d (216323_x_at), score: -0.63 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.53 UBA7ubiquitin-like modifier activating enzyme 7 (203281_s_at), score: 0.88 VAMP1vesicle-associated membrane protein 1 (synaptobrevin 1) (213326_at), score: 0.87 VCAM1vascular cell adhesion molecule 1 (203868_s_at), score: 0.74 VGLL3vestigial like 3 (Drosophila) (220327_at), score: 0.56 VPS11vacuolar protein sorting 11 homolog (S. cerevisiae) (203292_s_at), score: 0.57 WDR19WD repeat domain 19 (220917_s_at), score: 0.6 WFS1Wolfram syndrome 1 (wolframin) (202908_at), score: 0.53 ZBED4zinc finger, BED-type containing 4 (204799_at), score: -0.6 ZMAT3zinc finger, matrin type 3 (219628_at), score: 0.58 ZNF277zinc finger protein 277 (218645_at), score: 0.62 ZNF551zinc finger protein 551 (211721_s_at), score: -0.55 ZNF592zinc finger protein 592 (204473_s_at), score: -0.55 ZXDCZXD family zinc finger C (218639_s_at), score: -0.54
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| H652_WBS.CEL | 17 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |