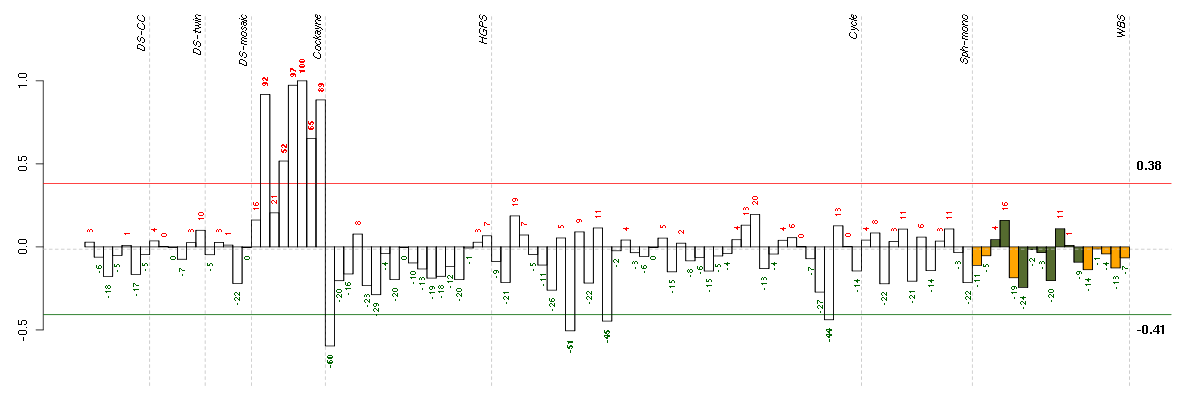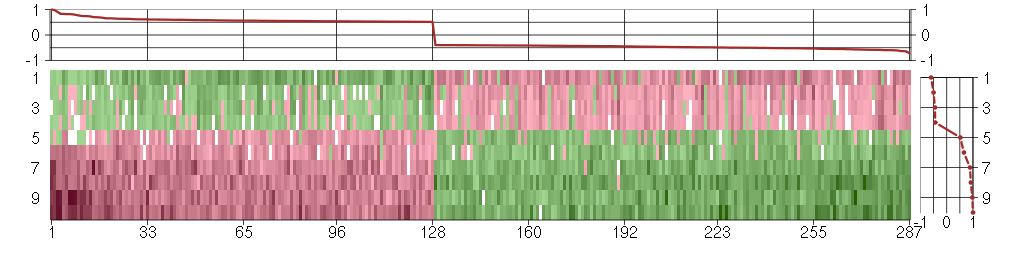

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
ATPase activity, coupled
Catalysis of the reaction: ATP + H2O = ADP + phosphate to directly drive some other reaction, for example ion transport across a membrane.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
nucleoside-triphosphatase activity
Catalysis of the reaction: a nucleoside triphosphate + H2O = nucleoside diphosphate + phosphate.
RNA-dependent ATPase activity
Catalysis of the reaction: ATP + H2O = ADP + phosphate, in the presence of RNA; drives another reaction.
ATPase activity
Catalysis of the reaction: ATP + H2O = ADP + phosphate. May or may not be coupled to another reaction.
pyrophosphatase activity
Catalysis of the hydrolysis of a pyrophosphate bond between two phosphate groups, leaving one phosphate on each of the two fragments.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on acid anhydrides
Catalysis of the hydrolysis of any acid anhydride.
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
Catalysis of the hydrolysis of any acid anhydride which contains phosphorus.
all
This term is the most general term possible
ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: 1 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: 0.7 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: 0.65 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.4 ADORA2Badenosine A2b receptor (205891_at), score: 0.53 AIM2absent in melanoma 2 (206513_at), score: 0.65 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: 0.63 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.52 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: 0.55 AOF2amine oxidase (flavin containing) domain 2 (212348_s_at), score: 0.53 APCadenomatous polyposis coli (203525_s_at), score: -0.44 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.57 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.56 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.56 ARHGAP1Rho GTPase activating protein 1 (202117_at), score: -0.45 ARNTLaryl hydrocarbon receptor nuclear translocator-like (209824_s_at), score: -0.51 ARSJarylsulfatase family, member J (219973_at), score: -0.44 ASXL2additional sex combs like 2 (Drosophila) (218659_at), score: 0.53 ATP11BATPase, class VI, type 11B (212536_at), score: -0.44 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: 0.64 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: -0.51 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.58 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: 0.63 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: 0.7 BMP6bone morphogenetic protein 6 (206176_at), score: -0.48 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: -0.52 C11orf17chromosome 11 open reading frame 17 (219953_s_at), score: 0.55 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.5 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: -0.46 C17orf91chromosome 17 open reading frame 91 (214696_at), score: -0.41 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.58 C4orf31chromosome 4 open reading frame 31 (219747_at), score: -0.56 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.51 C5orf30chromosome 5 open reading frame 30 (221823_at), score: -0.54 CA3carbonic anhydrase III, muscle specific (204865_at), score: 0.6 CARD10caspase recruitment domain family, member 10 (210026_s_at), score: 0.58 CCNL2cyclin L2 (221427_s_at), score: 0.56 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.56 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: 0.56 CDC2L2cell division cycle 2-like 2 (PITSLRE proteins) (212401_s_at), score: 0.52 CDKN2Dcyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) (210240_s_at), score: 0.52 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: -0.63 COL11A1collagen, type XI, alpha 1 (37892_at), score: -0.4 COMPcartilage oligomeric matrix protein (205713_s_at), score: -0.53 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: -0.46 CRTAPcartilage associated protein (201380_at), score: -0.57 CRYABcrystallin, alpha B (209283_at), score: -0.53 CTBP1C-terminal binding protein 1 (213979_s_at), score: 0.57 CTNNBL1catenin, beta like 1 (221021_s_at), score: 0.53 CUX1cut-like homeobox 1 (214743_at), score: -0.47 CYBAcytochrome b-245, alpha polypeptide (203028_s_at), score: -0.44 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: -0.4 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: 0.83 DDX3XDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, X-linked (201211_s_at), score: 0.6 DDX3YDEAD (Asp-Glu-Ala-Asp) box polypeptide 3, Y-linked (205000_at), score: -0.48 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: -0.42 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.54 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: 0.53 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: 0.55 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: 0.74 DIS3DIS3 mitotic control homolog (S. cerevisiae) (218362_s_at), score: -0.46 DLSTPdihydrolipoamide S-succinyltransferase pseudogene (E2 component of 2-oxo-glutarate complex) (215210_s_at), score: -0.45 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.56 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.54 ECSITECSIT homolog (Drosophila) (218225_at), score: 0.56 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.4 EDNRAendothelin receptor type A (204464_s_at), score: -0.4 EIF1AYeukaryotic translation initiation factor 1A, Y-linked (204409_s_at), score: -0.5 EIF4Beukaryotic translation initiation factor 4B (219599_at), score: 0.52 EPB41L2erythrocyte membrane protein band 4.1-like 2 (201718_s_at), score: 0.52 EPHB2EPH receptor B2 (209589_s_at), score: 0.65 ERO1LERO1-like (S. cerevisiae) (218498_s_at), score: -0.42 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.42 ETS1v-ets erythroblastosis virus E26 oncogene homolog 1 (avian) (214447_at), score: 0.53 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: -0.51 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.5 FAM169Afamily with sequence similarity 169, member A (213954_at), score: 0.68 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.49 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: -0.58 FBN2fibrillin 2 (203184_at), score: -0.5 FBXW7F-box and WD repeat domain containing 7 (218751_s_at), score: -0.41 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: -0.55 FLJ11151hypothetical protein FLJ11151 (218610_s_at), score: -0.42 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.55 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.45 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.52 GAGE4G antigen 4 (207086_x_at), score: 0.91 GAGE6G antigen 6 (208155_x_at), score: 0.83 GATA3GATA binding protein 3 (209604_s_at), score: 0.73 GBP1guanylate binding protein 1, interferon-inducible, 67kDa (202269_x_at), score: 0.54 GHRgrowth hormone receptor (205498_at), score: -0.59 GLE1GLE1 RNA export mediator homolog (yeast) (206920_s_at), score: 0.58 GLSglutaminase (203159_at), score: -0.49 GLTPglycolipid transfer protein (219267_at), score: -0.56 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: 0.55 GMPRguanosine monophosphate reductase (204187_at), score: 0.54 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.46 GSPT2G1 to S phase transition 2 (205541_s_at), score: -0.46 GYS1glycogen synthase 1 (muscle) (201673_s_at), score: -0.4 H1F0H1 histone family, member 0 (208886_at), score: 0.62 HAS2hyaluronan synthase 2 (206432_at), score: -0.55 HEPHhephaestin (203903_s_at), score: -0.41 HERC5hect domain and RLD 5 (219863_at), score: 0.74 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: -0.6 HIPK1homeodomain interacting protein kinase 1 (212291_at), score: 0.53 HIST1H4Chistone cluster 1, H4c (205967_at), score: -0.52 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: -0.52 HNRNPCheterogeneous nuclear ribonucleoprotein C (C1/C2) (200751_s_at), score: 0.59 HNRNPH1heterogeneous nuclear ribonucleoprotein H1 (H) (213470_s_at), score: 0.54 HRASLSHRAS-like suppressor (219983_at), score: 0.52 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.6 HSPB2heat shock 27kDa protein 2 (205824_at), score: -0.41 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: -0.61 IFI16interferon, gamma-inducible protein 16 (208965_s_at), score: 0.6 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.61 IGFBP3insulin-like growth factor binding protein 3 (212143_s_at), score: -0.55 IKIK cytokine, down-regulator of HLA II (200066_at), score: 0.52 IL11interleukin 11 (206924_at), score: -0.4 IL21Rinterleukin 21 receptor (219971_at), score: -0.43 IRS2insulin receptor substrate 2 (209185_s_at), score: -0.49 ISLRimmunoglobulin superfamily containing leucine-rich repeat (207191_s_at), score: -0.5 ITGBL1integrin, beta-like 1 (with EGF-like repeat domains) (214927_at), score: -0.43 JARID1Djumonji, AT rich interactive domain 1D (206700_s_at), score: -0.48 KCNS3potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 (205968_at), score: -0.4 KIAA0355KIAA0355 (203288_at), score: -0.43 KIAA0495KIAA0495 (213340_s_at), score: -0.58 KIF13Bkinesin family member 13B (202962_at), score: 0.55 KIF3Akinesin family member 3A (213623_at), score: 0.55 KIFC1kinesin family member C1 (209680_s_at), score: 0.58 KRT14keratin 14 (209351_at), score: -0.41 KRT19keratin 19 (201650_at), score: -0.4 LAMB3laminin, beta 3 (209270_at), score: 0.61 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: -0.4 LIMK2LIM domain kinase 2 (202193_at), score: -0.43 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: -0.45 LOC100132430hypothetical LOC100132430 (214182_at), score: 0.52 LOC151162hypothetical LOC151162 (212098_at), score: -0.44 LOC388796hypothetical LOC388796 (65588_at), score: 0.52 LOC399491LOC399491 protein (214035_x_at), score: 0.55 LOC728855hypothetical LOC728855 (222001_x_at), score: 0.57 LRIG2leucine-rich repeats and immunoglobulin-like domains 2 (205953_at), score: 0.53 LRRC2leucine rich repeat containing 2 (219949_at), score: -0.48 LRRFIP1leucine rich repeat (in FLII) interacting protein 1 (201861_s_at), score: -0.48 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.47 M6PRmannose-6-phosphate receptor (cation dependent) (200900_s_at), score: 0.6 MALLmal, T-cell differentiation protein-like (209373_at), score: -0.41 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: -0.58 MAP3K3mitogen-activated protein kinase kinase kinase 3 (203514_at), score: -0.4 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: -0.52 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: 0.77 MBOAT2membrane bound O-acyltransferase domain containing 2 (213288_at), score: -0.52 MED6mediator complex subunit 6 (207079_s_at), score: 0.54 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.52 MFAP5microfibrillar associated protein 5 (213764_s_at), score: -0.58 MGEA5meningioma expressed antigen 5 (hyaluronidase) (200898_s_at), score: 0.52 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: 0.52 MREGmelanoregulin (219648_at), score: -0.48 MRFAP1L1Morf4 family associated protein 1-like 1 (212199_at), score: 0.54 MT1Mmetallothionein 1M (217546_at), score: -0.53 MT1P3metallothionein 1 pseudogene 3 (221953_s_at), score: -0.48 MT1Xmetallothionein 1X (204326_x_at), score: -0.42 MYO10myosin X (201976_s_at), score: -0.51 MYO1Cmyosin IC (214656_x_at), score: -0.72 MYO1Dmyosin ID (212338_at), score: -0.43 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: 0.57 NBASneuroblastoma amplified sequence (202926_at), score: -0.42 NCALDneurocalcin delta (211685_s_at), score: -0.53 NCR2natural cytotoxicity triggering receptor 2 (217045_x_at), score: -0.47 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: 0.64 NDNnecdin homolog (mouse) (209550_at), score: -0.42 NEFLneurofilament, light polypeptide (221805_at), score: 0.55 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: -0.44 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: 0.56 NMIN-myc (and STAT) interactor (203964_at), score: 0.54 NMUneuromedin U (206023_at), score: 0.55 NOL12nucleolar protein 12 (219324_at), score: 0.54 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: 0.59 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: 0.66 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.54 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: 0.98 ODZ4odz, odd Oz/ten-m homolog 4 (Drosophila) (213273_at), score: -0.41 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.58 PARP4poly (ADP-ribose) polymerase family, member 4 (202239_at), score: 0.56 PCDH9protocadherin 9 (219737_s_at), score: -0.44 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.4 PDE10Aphosphodiesterase 10A (205501_at), score: 0.54 PDPK13-phosphoinositide dependent protein kinase-1 (204524_at), score: -0.57 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: 0.69 PODXLpodocalyxin-like (201578_at), score: -0.5 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: 0.56 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: -0.41 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: -0.41 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: -0.41 PRKCHprotein kinase C, eta (218764_at), score: 0.55 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.61 PTHLHparathyroid hormone-like hormone (211756_at), score: -0.6 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.53 PXDNperoxidasin homolog (Drosophila) (212013_at), score: -0.47 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: -0.46 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.41 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: 0.63 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: -0.5 RHODras homolog gene family, member D (31846_at), score: -0.4 RNF19Aring finger protein 19A (220483_s_at), score: 0.53 RPL31ribosomal protein L31 (221593_s_at), score: -0.43 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: 0.53 RPS4Y1ribosomal protein S4, Y-linked 1 (201909_at), score: -0.43 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: 0.57 SC65synaptonemal complex protein SC65 (204078_at), score: -0.4 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.52 SEC16ASEC16 homolog A (S. cerevisiae) (215696_s_at), score: -0.41 SEPHS1selenophosphate synthetase 1 (208939_at), score: 0.51 SFPQsplicing factor proline/glutamine-rich (polypyrimidine tract binding protein associated) (201585_s_at), score: 0.53 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.56 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: -0.5 SHANK2SH3 and multiple ankyrin repeat domains 2 (213307_at), score: -0.41 SIDT2SID1 transmembrane family, member 2 (56256_at), score: -0.48 SLC10A3solute carrier family 10 (sodium/bile acid cotransporter family), member 3 (204928_s_at), score: -0.41 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: -0.59 SLC1A1solute carrier family 1 (neuronal/epithelial high affinity glutamate transporter, system Xag), member 1 (213664_at), score: -0.46 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.47 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: -0.43 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.55 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: -0.48 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.65 SLC7A1solute carrier family 7 (cationic amino acid transporter, y+ system), member 1 (212295_s_at), score: -0.41 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: 0.6 SMAD3SMAD family member 3 (218284_at), score: 0.51 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: 0.62 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: 0.6 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.82 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: -0.52 SPINT2serine peptidase inhibitor, Kunitz type, 2 (210715_s_at), score: -0.42 SS18synovial sarcoma translocation, chromosome 18 (216684_s_at), score: 0.53 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: -0.63 SSRP1structure specific recognition protein 1 (200956_s_at), score: 0.52 SUPT16Hsuppressor of Ty 16 homolog (S. cerevisiae) (217815_at), score: 0.58 SYTL2synaptotagmin-like 2 (220613_s_at), score: 0.56 TACSTD2tumor-associated calcium signal transducer 2 (202286_s_at), score: -0.5 TBC1D3TBC1 domain family, member 3 (209403_at), score: 0.54 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.56 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: 0.75 TEKTEK tyrosine kinase, endothelial (206702_at), score: -0.47 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: 0.6 TGFAtransforming growth factor, alpha (205016_at), score: -0.44 TGIF1TGFB-induced factor homeobox 1 (203313_s_at), score: 0.57 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.58 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: -0.43 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.47 TMEM204transmembrane protein 204 (219315_s_at), score: -0.54 TMEM47transmembrane protein 47 (209656_s_at), score: -0.49 TMEM51transmembrane protein 51 (218815_s_at), score: 0.61 TMEM97transmembrane protein 97 (212279_at), score: 0.59 TMOD1tropomodulin 1 (203661_s_at), score: 0.52 TNS1tensin 1 (221748_s_at), score: -0.56 TOB1transducer of ERBB2, 1 (202704_at), score: -0.54 TRIM16tripartite motif-containing 16 (204341_at), score: 0.61 TSNtranslin (201504_s_at), score: 0.54 TSPAN13tetraspanin 13 (217979_at), score: -0.55 TSPAN31tetraspanin 31 (203227_s_at), score: -0.4 TSPYL1TSPY-like 1 (221493_at), score: -0.5 TSPYL5TSPY-like 5 (213122_at), score: -0.48 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: -0.41 TXNRD1thioredoxin reductase 1 (201266_at), score: 0.52 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.57 UBE2D4ubiquitin-conjugating enzyme E2D 4 (putative) (218837_s_at), score: -0.45 UBE2Hubiquitin-conjugating enzyme E2H (UBC8 homolog, yeast) (221962_s_at), score: 0.54 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.55 UBL3ubiquitin-like 3 (201535_at), score: -0.59 UCHL1ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) (201387_s_at), score: -0.47 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: 0.81 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: 0.82 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: 0.79 UROSuroporphyrinogen III synthase (203031_s_at), score: -0.43 USP18ubiquitin specific peptidase 18 (219211_at), score: 0.59 USP25ubiquitin specific peptidase 25 (220419_s_at), score: -0.43 UTP14CUTP14, U3 small nucleolar ribonucleoprotein, homolog C (yeast) (203614_at), score: -0.42 VEGFCvascular endothelial growth factor C (209946_at), score: -0.4 VLDLRvery low density lipoprotein receptor (209822_s_at), score: -0.4 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.61 WDR48WD repeat domain 48 (222157_s_at), score: 0.59 WT1Wilms tumor 1 (206067_s_at), score: 0.61 XYLT1xylosyltransferase I (213725_x_at), score: -0.42 ZNF227zinc finger protein 227 (217403_s_at), score: 0.55 ZNF281zinc finger protein 281 (218401_s_at), score: -0.52 ZNF415zinc finger protein 415 (205514_at), score: -0.48 ZNF654zinc finger protein 654 (219239_s_at), score: -0.44
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949704.cel | 4 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949854.cel | 7 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |