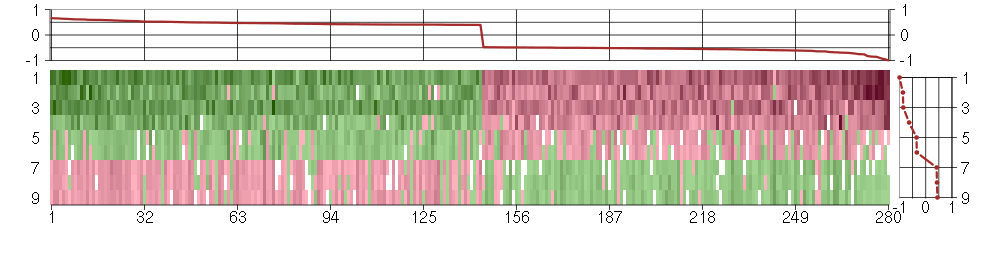

Under-expression is coded with green,
over-expression with red color.

biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to xenobiotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a xenobiotic compound stimulus. Xenobiotic compounds are compounds foreign to living organisms.
response to chemical stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a chemical stimulus.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
extracellular matrix part
Any constituent part of the extracellular matrix, the structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as often seen in plants).

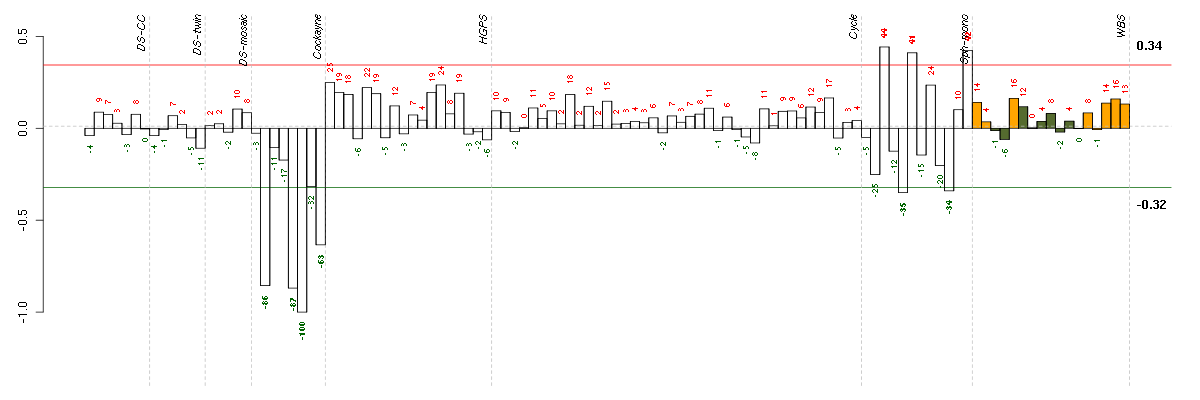
A2Malpha-2-macroglobulin (217757_at), score: -0.52 ABCB1ATP-binding cassette, sub-family B (MDR/TAP), member 1 (209993_at), score: -1 ABCB4ATP-binding cassette, sub-family B (MDR/TAP), member 4 (207819_s_at), score: -0.69 ABHD14Aabhydrolase domain containing 14A (210006_at), score: 0.44 ACIN1apoptotic chromatin condensation inducer 1 (201715_s_at), score: -0.49 ADCY7adenylate cyclase 7 (203741_s_at), score: 0.41 ADORA2Badenosine A2b receptor (205891_at), score: -0.55 AGPSalkylglycerone phosphate synthase (205401_at), score: -0.55 AIM2absent in melanoma 2 (206513_at), score: -0.61 AKAP8LA kinase (PRKA) anchor protein 8-like (218064_s_at), score: -0.57 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.66 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (201612_at), score: 0.42 ANK3ankyrin 3, node of Ranvier (ankyrin G) (206385_s_at), score: -0.54 ANP32Aacidic (leucine-rich) nuclear phosphoprotein 32 family, member A (201043_s_at), score: -0.54 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: 0.49 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: -0.49 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: -0.59 APOL3apolipoprotein L, 3 (221087_s_at), score: -0.5 ARMCX3armadillo repeat containing, X-linked 3 (217858_s_at), score: 0.41 ARSJarylsulfatase family, member J (219973_at), score: 0.5 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: -0.48 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: 0.41 BAZ2Abromodomain adjacent to zinc finger domain, 2A (201353_s_at), score: -0.51 BHMT2betaine-homocysteine methyltransferase 2 (219902_at), score: -0.53 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: 0.52 C17orf70chromosome 17 open reading frame 70 (221800_s_at), score: 0.41 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.41 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.41 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: 0.56 C4orf31chromosome 4 open reading frame 31 (219747_at), score: 0.45 C5orf30chromosome 5 open reading frame 30 (221823_at), score: 0.62 CA3carbonic anhydrase III, muscle specific (204865_at), score: -0.59 CAV2caveolin 2 (203323_at), score: 0.43 CCNG1cyclin G1 (208796_s_at), score: 0.51 CCNL2cyclin L2 (221427_s_at), score: -0.5 CD24CD24 molecule (209771_x_at), score: -0.61 CD248CD248 molecule, endosialin (219025_at), score: 0.57 CD81CD81 molecule (200675_at), score: 0.46 CDC25Ccell division cycle 25 homolog C (S. pombe) (205167_s_at), score: -0.51 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: 0.58 CDR2Lcerebellar degeneration-related protein 2-like (213230_at), score: 0.44 COL11A1collagen, type XI, alpha 1 (37892_at), score: 0.59 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.53 COPZ2coatomer protein complex, subunit zeta 2 (219561_at), score: 0.46 CRTAPcartilage associated protein (201380_at), score: 0.59 CRYABcrystallin, alpha B (209283_at), score: 0.54 CTNNBL1catenin, beta like 1 (221021_s_at), score: -0.61 CTSKcathepsin K (202450_s_at), score: 0.49 DBF4DBF4 homolog (S. cerevisiae) (204244_s_at), score: -0.49 DCNdecorin (211896_s_at), score: 0.41 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: 0.47 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (208151_x_at), score: -0.61 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: -0.68 DHX38DEAH (Asp-Glu-Ala-His) box polypeptide 38 (209178_at), score: -0.6 DHX9DEAH (Asp-Glu-Ala-His) box polypeptide 9 (212105_s_at), score: -0.49 DIO2deiodinase, iodothyronine, type II (203699_s_at), score: -0.85 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.51 DNASE1L1deoxyribonuclease I-like 1 (203912_s_at), score: 0.41 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: -0.51 DSPdesmoplakin (200606_at), score: 0.41 DUSP14dual specificity phosphatase 14 (203367_at), score: 0.52 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.41 EPHB2EPH receptor B2 (209589_s_at), score: -0.72 EWSR1Ewing sarcoma breakpoint region 1 (209214_s_at), score: -0.53 FAM105Afamily with sequence similarity 105, member A (219694_at), score: -0.62 FAM108A1family with sequence similarity 108, member A1 (221267_s_at), score: 0.41 FAM169Afamily with sequence similarity 169, member A (213954_at), score: -0.64 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.41 FARP1FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) (201911_s_at), score: 0.47 FAT1FAT tumor suppressor homolog 1 (Drosophila) (201579_at), score: 0.65 FBN2fibrillin 2 (203184_at), score: 0.48 FKBP4FK506 binding protein 4, 59kDa (200895_s_at), score: -0.51 FKBP9FK506 binding protein 9, 63 kDa (212169_at), score: 0.63 FLRT2fibronectin leucine rich transmembrane protein 2 (204359_at), score: 0.4 FOLR3folate receptor 3 (gamma) (206371_at), score: 0.44 FOXG1forkhead box G1 (206018_at), score: -0.54 FOXL2forkhead box L2 (220102_at), score: -0.49 FUSfusion (involved in t(12;16) in malignant liposarcoma) (217370_x_at), score: -0.5 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: 0.4 GAGE4G antigen 4 (207086_x_at), score: -0.93 GAGE6G antigen 6 (208155_x_at), score: -0.89 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: 0.47 GAS1growth arrest-specific 1 (204457_s_at), score: 0.42 GAS6growth arrest-specific 6 (202177_at), score: 0.49 GATA3GATA binding protein 3 (209604_s_at), score: -0.76 GCLCglutamate-cysteine ligase, catalytic subunit (202923_s_at), score: -0.49 GHRgrowth hormone receptor (205498_at), score: 0.47 GLSglutaminase (203159_at), score: 0.59 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: 0.41 GLTPglycolipid transfer protein (219267_at), score: 0.47 GMDSGDP-mannose 4,6-dehydratase (204875_s_at), score: -0.49 GNL3guanine nucleotide binding protein-like 3 (nucleolar) (217850_at), score: -0.52 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 0.46 GSRglutathione reductase (205770_at), score: -0.5 GTF2H4general transcription factor IIH, polypeptide 4, 52kDa (203577_at), score: -0.51 HAS2hyaluronan synthase 2 (206432_at), score: 0.53 HERC5hect domain and RLD 5 (219863_at), score: -0.75 HGSNATheparan-alpha-glucosaminide N-acetyltransferase (218017_s_at), score: 0.61 HIPK2homeodomain interacting protein kinase 2 (219028_at), score: -0.48 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.49 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: -0.5 HRASLSHRAS-like suppressor (219983_at), score: -0.55 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: 0.42 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: -0.56 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.4 HSPB6heat shock protein, alpha-crystallin-related, B6 (214767_s_at), score: 0.41 HSPB7heat shock 27kDa protein family, member 7 (cardiovascular) (218934_s_at), score: 0.52 HSPG2heparan sulfate proteoglycan 2 (201655_s_at), score: 0.42 IFI44interferon-induced protein 44 (214453_s_at), score: -0.49 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: -0.49 IGF2BP3insulin-like growth factor 2 mRNA binding protein 3 (203819_s_at), score: -0.49 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.56 IKIK cytokine, down-regulator of HLA II (200066_at), score: -0.55 IL11RAinterleukin 11 receptor, alpha (204773_at), score: 0.42 IQWD1IQ motif and WD repeats 1 (217908_s_at), score: 0.41 IRS2insulin receptor substrate 2 (209185_s_at), score: 0.63 KCNK2potassium channel, subfamily K, member 2 (210261_at), score: 0.41 KCNS3potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 (205968_at), score: 0.4 KIAA0495KIAA0495 (213340_s_at), score: 0.41 KIAA0802KIAA0802 (213358_at), score: 0.44 KIAA0895KIAA0895 (213424_at), score: -0.5 KIAA1128KIAA1128 (209379_s_at), score: 0.41 KIFC1kinesin family member C1 (209680_s_at), score: -0.56 KRT34keratin 34 (206969_at), score: 0.42 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.59 LAMB3laminin, beta 3 (209270_at), score: -0.6 LBHlimb bud and heart development homolog (mouse) (221011_s_at), score: 0.49 LDOC1leucine zipper, down-regulated in cancer 1 (204454_at), score: 0.5 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.68 LGMNlegumain (201212_at), score: 0.42 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: 0.53 LMO7LIM domain 7 (202674_s_at), score: 0.41 LOC388796hypothetical LOC388796 (65588_at), score: -0.55 LOC728855hypothetical LOC728855 (222001_x_at), score: -0.7 LRP10low density lipoprotein receptor-related protein 10 (201412_at), score: 0.41 LRRC15leucine rich repeat containing 15 (213909_at), score: 0.4 LRRC2leucine rich repeat containing 2 (219949_at), score: 0.61 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: 0.61 LUMlumican (201744_s_at), score: 0.46 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: -0.56 MAN2A1mannosidase, alpha, class 2A, member 1 (205105_at), score: 0.61 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: 0.47 MARK1MAP/microtubule affinity-regulating kinase 1 (221047_s_at), score: -0.64 MED6mediator complex subunit 6 (207079_s_at), score: -0.51 MFAP5microfibrillar associated protein 5 (213764_s_at), score: 0.59 MITFmicrophthalmia-associated transcription factor (207233_s_at), score: -0.54 MOXD1monooxygenase, DBH-like 1 (209708_at), score: 0.57 MRC2mannose receptor, C type 2 (37408_at), score: 0.49 MRTO4mRNA turnover 4 homolog (S. cerevisiae) (220688_s_at), score: -0.49 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: -0.53 MXRA5matrix-remodelling associated 5 (209596_at), score: 0.49 MXRA8matrix-remodelling associated 8 (213422_s_at), score: 0.52 MYO1Cmyosin IC (214656_x_at), score: 0.49 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: -0.5 NCK2NCK adaptor protein 2 (203315_at), score: -0.51 NDNnecdin homolog (mouse) (209550_at), score: 0.47 NEFLneurofilament, light polypeptide (221805_at), score: -0.69 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: 0.52 NFASCneurofascin homolog (chicken) (213438_at), score: 0.52 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: -0.59 NIP7nuclear import 7 homolog (S. cerevisiae) (219031_s_at), score: -0.49 NMUneuromedin U (206023_at), score: -0.6 NOC2Lnucleolar complex associated 2 homolog (S. cerevisiae) (202115_s_at), score: -0.58 NOL12nucleolar protein 12 (219324_at), score: -0.62 NONOnon-POU domain containing, octamer-binding (208698_s_at), score: -0.6 NRGNneurogranin (protein kinase C substrate, RC3) (204081_at), score: -0.64 OASL2'-5'-oligoadenylate synthetase-like (210797_s_at), score: -0.96 ODF2outer dense fiber of sperm tails 2 (210415_s_at), score: -0.5 OLFML3olfactomedin-like 3 (218162_at), score: 0.4 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (207543_s_at), score: 0.45 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: 0.52 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.7 PDE10Aphosphodiesterase 10A (205501_at), score: -0.55 PDGFCplatelet derived growth factor C (218718_at), score: 0.55 PDZRN3PDZ domain containing ring finger 3 (212915_at), score: 0.4 PHF11PHD finger protein 11 (221816_s_at), score: 0.43 PILRBpaired immunoglobin-like type 2 receptor beta (220954_s_at), score: -0.64 PIRpirin (iron-binding nuclear protein) (207469_s_at), score: -0.49 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: 0.43 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: -0.48 PODXLpodocalyxin-like (201578_at), score: 0.4 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.48 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: -0.51 PPM2Cprotein phosphatase 2C, magnesium-dependent, catalytic subunit (218273_s_at), score: -0.5 PPP1R3Cprotein phosphatase 1, regulatory (inhibitor) subunit 3C (204284_at), score: 0.45 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: 0.44 PQBP1polyglutamine binding protein 1 (214527_s_at), score: -0.62 PSG2pregnancy specific beta-1-glycoprotein 2 (208134_x_at), score: 0.44 PSG5pregnancy specific beta-1-glycoprotein 5 (204830_x_at), score: 0.4 PSG9pregnancy specific beta-1-glycoprotein 9 (209594_x_at), score: 0.4 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: -0.6 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: -0.53 PTGER3prostaglandin E receptor 3 (subtype EP3) (213933_at), score: 0.49 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.55 QRSL1glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 (218949_s_at), score: -0.48 R3HCC1R3H domain and coiled-coil containing 1 (212866_at), score: -0.56 R3HDM2R3H domain containing 2 (203831_at), score: 0.42 RAB27ARAB27A, member RAS oncogene family (210951_x_at), score: -0.49 RAB7L1RAB7, member RAS oncogene family-like 1 (218700_s_at), score: -0.54 RABGAP1LRAB GTPase activating protein 1-like (213982_s_at), score: 0.5 RALYRNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) (201271_s_at), score: -0.51 RARRES3retinoic acid receptor responder (tazarotene induced) 3 (204070_at), score: -0.52 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: 0.6 REEP1receptor accessory protein 1 (204364_s_at), score: -0.53 RGNregucalcin (senescence marker protein-30) (210751_s_at), score: 0.41 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: -0.59 RHOBTB1Rho-related BTB domain containing 1 (212651_at), score: 0.41 RHOQras homolog gene family, member Q (212119_at), score: 0.46 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: -0.54 RSL1D1ribosomal L1 domain containing 1 (212018_s_at), score: -0.52 SAFB2scaffold attachment factor B2 (32099_at), score: -0.52 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: 0.55 SART3squamous cell carcinoma antigen recognized by T cells 3 (209127_s_at), score: -0.52 SC65synaptonemal complex protein SC65 (204078_at), score: 0.44 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.58 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.4 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.41 SF4splicing factor 4 (215004_s_at), score: -0.53 SGCDsarcoglycan, delta (35kDa dystrophin-associated glycoprotein) (213543_at), score: 0.46 SH3YL1SH3 domain containing, Ysc84-like 1 (S. cerevisiae) (204019_s_at), score: 0.43 SHFM1split hand/foot malformation (ectrodactyly) type 1 (202276_at), score: -0.5 SIDT2SID1 transmembrane family, member 2 (56256_at), score: 0.45 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: -0.52 SLC16A1solute carrier family 16, member 1 (monocarboxylic acid transporter 1) (209900_s_at), score: 0.4 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: 0.46 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.51 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: 0.43 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: 0.52 SLC7A11solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 (217678_at), score: -0.58 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (220368_s_at), score: -0.56 SMG1SMG1 homolog, phosphatidylinositol 3-kinase-related kinase (C. elegans) (210057_at), score: -0.53 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: 0.46 SNAPC1small nuclear RNA activating complex, polypeptide 1, 43kDa (205443_at), score: -0.48 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: -0.67 SPARCsecreted protein, acidic, cysteine-rich (osteonectin) (212667_at), score: 0.47 SSNA1Sjogren syndrome nuclear autoantigen 1 (210378_s_at), score: -0.53 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: 0.41 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (217790_s_at), score: 0.5 SSRP1structure specific recognition protein 1 (200956_s_at), score: -0.57 STOML2stomatin (EPB72)-like 2 (215416_s_at), score: -0.49 STSsteroid sulfatase (microsomal), isozyme S (203767_s_at), score: 0.47 STX2syntaxin 2 (213434_at), score: 0.43 STXBP6syntaxin binding protein 6 (amisyn) (220994_s_at), score: 0.41 SYTL2synaptotagmin-like 2 (220613_s_at), score: -0.55 TBC1D3TBC1 domain family, member 3 (209403_at), score: -0.54 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: -0.58 TCEAL2transcription elongation factor A (SII)-like 2 (211276_at), score: -0.75 TEKTEK tyrosine kinase, endothelial (206702_at), score: 0.54 TERF1telomeric repeat binding factor (NIMA-interacting) 1 (203448_s_at), score: -0.57 TESK1testis-specific kinase 1 (204106_at), score: 0.42 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.64 TKTtransketolase (208699_x_at), score: -0.54 TMEM204transmembrane protein 204 (219315_s_at), score: 0.48 TMEM47transmembrane protein 47 (209656_s_at), score: 0.64 TMOD1tropomodulin 1 (203661_s_at), score: -0.59 TNS1tensin 1 (221748_s_at), score: 0.65 TOB1transducer of ERBB2, 1 (202704_at), score: 0.58 TOE1target of EGR1, member 1 (nuclear) (204080_at), score: -0.49 TPD52tumor protein D52 (201690_s_at), score: -0.49 TRIM16tripartite motif-containing 16 (204341_at), score: -0.56 TRIM8tripartite motif-containing 8 (221012_s_at), score: 0.44 TSPAN13tetraspanin 13 (217979_at), score: 0.59 TSPYL1TSPY-like 1 (221493_at), score: 0.48 TSPYL5TSPY-like 5 (213122_at), score: 0.56 TTTY15testis-specific transcript, Y-linked 15 (214983_at), score: 0.39 TUBG2tubulin, gamma 2 (203894_at), score: 0.42 TYRP1tyrosinase-related protein 1 (205694_at), score: -0.63 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: 0.52 UBL3ubiquitin-like 3 (201535_at), score: 0.66 UGT1A1UDP glucuronosyltransferase 1 family, polypeptide A1 (207126_x_at), score: -0.85 UGT1A6UDP glucuronosyltransferase 1 family, polypeptide A6 (206094_x_at), score: -0.83 UGT1A9UDP glucuronosyltransferase 1 family, polypeptide A9 (204532_x_at), score: -0.84 URODuroporphyrinogen decarboxylase (208971_at), score: -0.48 USP18ubiquitin specific peptidase 18 (219211_at), score: -0.51 UXS1UDP-glucuronate decarboxylase 1 (219675_s_at), score: 0.41 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: -0.56 WDR74WD repeat domain 74 (221712_s_at), score: -0.58 WT1Wilms tumor 1 (206067_s_at), score: -0.73 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.48 ZNF804Azinc finger protein 804A (215767_at), score: -0.51
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-3407-raw-cel-1437949750.cel | 6 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-3407-raw-cel-1437949938.cel | 8 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |