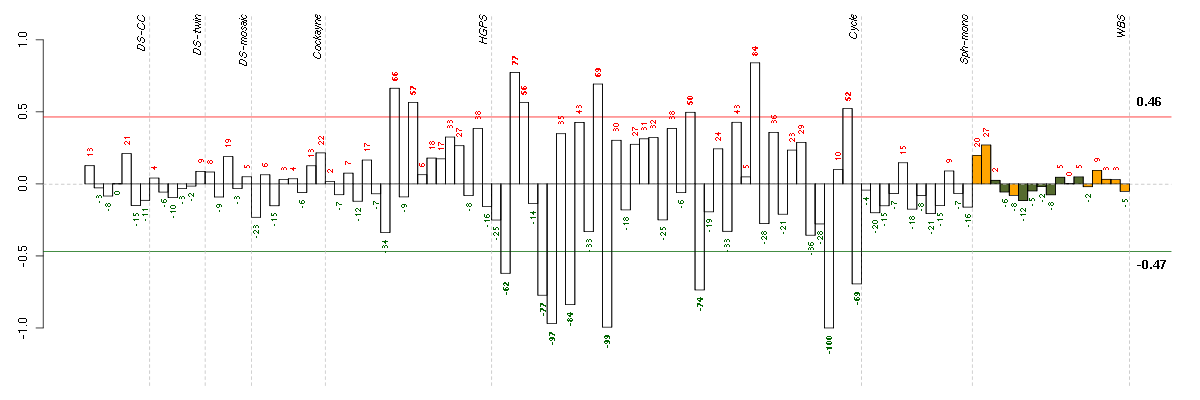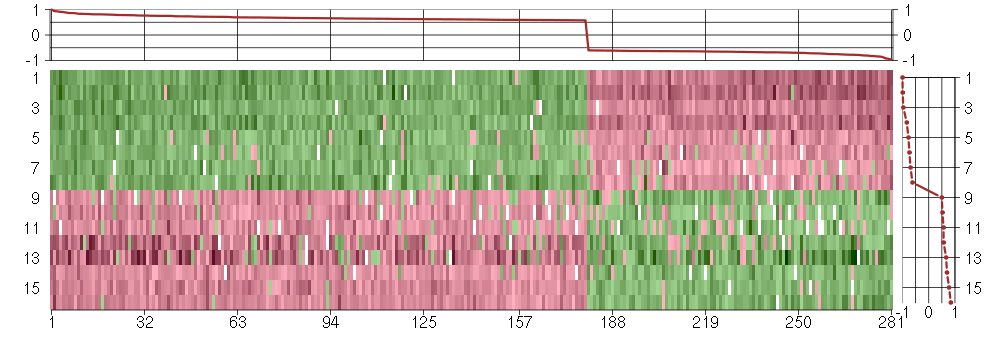

Under-expression is coded with green,
over-expression with red color.



| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05050 | 2.844e-02 | 0.3852 | 4 | 14 | Dentatorubropallidoluysian atrophy (DRPLA) |
AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.62 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.71 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.68 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.68 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.76 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.62 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.74 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.65 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.65 ARSAarylsulfatase A (204443_at), score: 0.62 ASF1BASF1 anti-silencing function 1 homolog B (S. cerevisiae) (218115_at), score: 0.61 ATF2activating transcription factor 2 (212984_at), score: -0.63 ATF5activating transcription factor 5 (204999_s_at), score: 0.61 ATN1atrophin 1 (40489_at), score: 0.75 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.75 AZI25-azacytidine induced 2 (218043_s_at), score: -0.63 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.73 BACE2beta-site APP-cleaving enzyme 2 (217867_x_at), score: 0.68 BAIAP2BAI1-associated protein 2 (209502_s_at), score: 0.72 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.64 BAT1HLA-B associated transcript 1 (200041_s_at), score: 0.58 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.78 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.64 BAT3HLA-B associated transcript 3 (213318_s_at), score: 0.58 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.81 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.65 C14orf104chromosome 14 open reading frame 104 (219166_at), score: -0.64 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.68 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.64 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.64 C19orf61chromosome 19 open reading frame 61 (221335_x_at), score: 0.67 C20orf117chromosome 20 open reading frame 117 (207711_at), score: 0.61 C20orf20chromosome 20 open reading frame 20 (218586_at), score: 0.59 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.81 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.72 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: -0.63 CABIN1calcineurin binding protein 1 (37652_at), score: 0.63 CALB2calbindin 2 (205428_s_at), score: -0.7 CAPGcapping protein (actin filament), gelsolin-like (201850_at), score: 0.59 CAPN5calpain 5 (205166_at), score: 0.62 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.72 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.67 CD248CD248 molecule, endosialin (219025_at), score: 0.6 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.72 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.63 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.86 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.59 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.69 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.77 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.66 CENPTcentromere protein T (218148_at), score: 0.61 CHMP5chromatin modifying protein 5 (218085_at), score: -0.74 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.61 CICcapicua homolog (Drosophila) (212784_at), score: 0.83 CITcitron (rho-interacting, serine/threonine kinase 21) (212801_at), score: 0.81 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.8 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.73 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.88 CORINcorin, serine peptidase (220356_at), score: -0.62 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.91 CScitrate synthase (208660_at), score: 0.75 CUX1cut-like homeobox 1 (214743_at), score: -0.62 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.59 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.63 DERL2Der1-like domain family, member 2 (218333_at), score: -0.61 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.59 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.82 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.64 DNM1dynamin 1 (215116_s_at), score: 0.59 DNM2dynamin 2 (202253_s_at), score: 0.61 DOPEY1dopey family member 1 (40612_at), score: -0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.62 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.61 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.62 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.61 EPHA4EPH receptor A4 (206114_at), score: -0.65 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.64 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.81 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.63 FAM62Afamily with sequence similarity 62 (C2 domain containing), member A (208858_s_at), score: 0.58 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: -0.6 FBXL11F-box and leucine-rich repeat protein 11 (208989_s_at), score: 0.6 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.69 FBXO17F-box protein 17 (220233_at), score: 0.59 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.63 FKRPfukutin related protein (219853_at), score: 0.64 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.81 FLOT1flotillin 1 (210142_x_at), score: 0.63 FOSL1FOS-like antigen 1 (204420_at), score: 0.68 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.58 FOXK2forkhead box K2 (203064_s_at), score: 0.85 FOXM1forkhead box M1 (202580_x_at), score: 0.66 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.71 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.65 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.67 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.64 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.66 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.72 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: -0.61 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.64 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.6 H1F0H1 histone family, member 0 (208886_at), score: 0.66 H1FXH1 histone family, member X (204805_s_at), score: 0.78 HAB1B1 for mucin (215778_x_at), score: -0.64 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.69 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.65 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.76 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.78 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.77 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.84 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.81 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: -0.65 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.75 ITGA1integrin, alpha 1 (214660_at), score: -0.75 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.64 JUNDjun D proto-oncogene (203751_x_at), score: 0.59 JUPjunction plakoglobin (201015_s_at), score: 0.65 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.94 KIAA0562KIAA0562 (204075_s_at), score: -0.62 KIAA1305KIAA1305 (220911_s_at), score: 0.59 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.7 KPNA6karyopherin alpha 6 (importin alpha 7) (212101_at), score: 0.59 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.73 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.65 LEPREL2leprecan-like 2 (204854_at), score: 0.61 LIG1ligase I, DNA, ATP-dependent (202726_at), score: 0.66 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.6 LMF2lipase maturation factor 2 (212682_s_at), score: 0.66 LMNB2lamin B2 (216952_s_at), score: 0.65 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.71 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.63 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.75 LOC391132similar to hCG2041276 (216177_at), score: -0.77 LOC399491LOC399491 protein (214035_x_at), score: 0.58 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.59 LOC90379hypothetical protein BC002926 (221849_s_at), score: 0.62 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.64 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.8 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.66 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.77 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.86 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.78 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.73 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.59 MAPKAPK3mitogen-activated protein kinase-activated protein kinase 3 (202788_at), score: 0.69 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: 0.76 MED15mediator complex subunit 15 (222175_s_at), score: 0.64 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.79 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.61 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.66 MLL4myeloid/lymphoid or mixed-lineage leukemia 4 (203419_at), score: 0.62 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.67 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.63 MYO1Bmyosin IB (212364_at), score: -0.69 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.82 NCKAP1NCK-associated protein 1 (217465_at), score: -0.7 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.59 NDPNorrie disease (pseudoglioma) (206022_at), score: 0.75 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.65 NEFLneurofilament, light polypeptide (221805_at), score: -0.64 NEFMneurofilament, medium polypeptide (205113_at), score: -0.96 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.58 NF1neurofibromin 1 (211094_s_at), score: 0.65 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.73 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.6 NLGN4Yneuroligin 4, Y-linked (207703_at), score: -0.61 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.79 NRXN3neurexin 3 (205795_at), score: -0.7 NUPL2nucleoporin like 2 (204003_s_at), score: -0.64 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.66 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.94 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.68 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.72 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.63 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.7 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.72 PARVBparvin, beta (204629_at), score: 0.75 PBX3pre-B-cell leukemia homeobox 3 (204082_at), score: 0.61 PCDH9protocadherin 9 (219737_s_at), score: -0.78 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.67 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.76 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.63 PCNTpericentrin (203660_s_at), score: 0.58 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.59 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.58 PHF7PHD finger protein 7 (215622_x_at), score: -0.67 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.82 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.64 PKN1protein kinase N1 (202161_at), score: 0.69 PLCD1phospholipase C, delta 1 (205125_at), score: 0.6 PLK1polo-like kinase 1 (Drosophila) (202240_at), score: 0.69 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.66 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.71 POLSpolymerase (DNA directed) sigma (202466_at), score: 0.73 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.78 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.6 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.7 PRKCDprotein kinase C, delta (202545_at), score: 0.65 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.83 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.63 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.66 PTTG3pituitary tumor-transforming 3 (208511_at), score: 0.59 PXNpaxillin (211823_s_at), score: 0.67 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.75 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.66 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: 0.64 RAG1AP1recombination activating gene 1 activating protein 1 (219125_s_at), score: -0.62 RBM15BRNA binding motif protein 15B (202689_at), score: 0.68 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.78 RNF11ring finger protein 11 (208924_at), score: -0.76 RNF220ring finger protein 220 (219988_s_at), score: 0.93 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.91 RPRD2regulation of nuclear pre-mRNA domain containing 2 (212553_at), score: 0.64 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.84 RPS25ribosomal protein S25 (200091_s_at), score: -0.66 RPS6KA4ribosomal protein S6 kinase, 90kDa, polypeptide 4 (204632_at), score: 0.61 SAP30LSAP30-like (219129_s_at), score: 0.69 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.69 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 1 SCARA3scavenger receptor class A, member 3 (219416_at), score: 0.62 SDC1syndecan 1 (201287_s_at), score: 0.62 SEC61A2Sec61 alpha 2 subunit (S. cerevisiae) (219499_at), score: 0.66 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.59 SF1splicing factor 1 (208313_s_at), score: 0.9 SF3A2splicing factor 3a, subunit 2, 66kDa (37462_i_at), score: 0.68 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.63 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.64 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.76 SLC43A3solute carrier family 43, member 3 (213113_s_at), score: 0.75 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.63 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.61 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.69 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.67 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (201794_s_at), score: 0.65 SNAP23synaptosomal-associated protein, 23kDa (214544_s_at), score: -0.61 SNAP25synaptosomal-associated protein, 25kDa (202508_s_at), score: 0.65 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.83 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.66 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.59 STIM1stromal interaction molecule 1 (202764_at), score: 0.71 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.71 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.75 SUV39H1suppressor of variegation 3-9 homolog 1 (Drosophila) (218619_s_at), score: 0.6 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.67 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.63 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.71 TBCDtubulin folding cofactor D (211052_s_at), score: 0.68 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.62 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.71 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.86 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.69 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.68 TMEM123transmembrane protein 123 (211967_at), score: -0.63 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.58 TMSB15Bthymosin beta 15B (214051_at), score: 0.64 TRIM21tripartite motif-containing 21 (204804_at), score: 0.68 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.58 TSKUtsukushin (218245_at), score: 0.64 TXLNAtaxilin alpha (212300_at), score: 0.78 UBE2Zubiquitin-conjugating enzyme E2Z (217750_s_at), score: 0.59 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.72 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.67 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.61 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.65 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.81 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.76 WDR42AWD repeat domain 42A (202249_s_at), score: 0.62 WDR6WD repeat domain 6 (217734_s_at), score: 0.83 WDR62WD repeat domain 62 (215218_s_at), score: 0.69 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.61 WRAP53WD repeat containing, antisense to TP53 (44563_at), score: 0.58 XAB2XPA binding protein 2 (218110_at), score: 0.68 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.73 YTHDC2YTH domain containing 2 (213077_at), score: -0.72 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.68 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.63 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.67 ZFYVE26zinc finger, FYVE domain containing 26 (37943_at), score: -0.68 ZNF281zinc finger protein 281 (218401_s_at), score: -0.62 ZNF318zinc finger protein 318 (203521_s_at), score: 0.69 ZNF536zinc finger protein 536 (206403_at), score: 0.61
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690344.cel | 10 | 5 | HGPS | hgu133a | none | GM00038C |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |