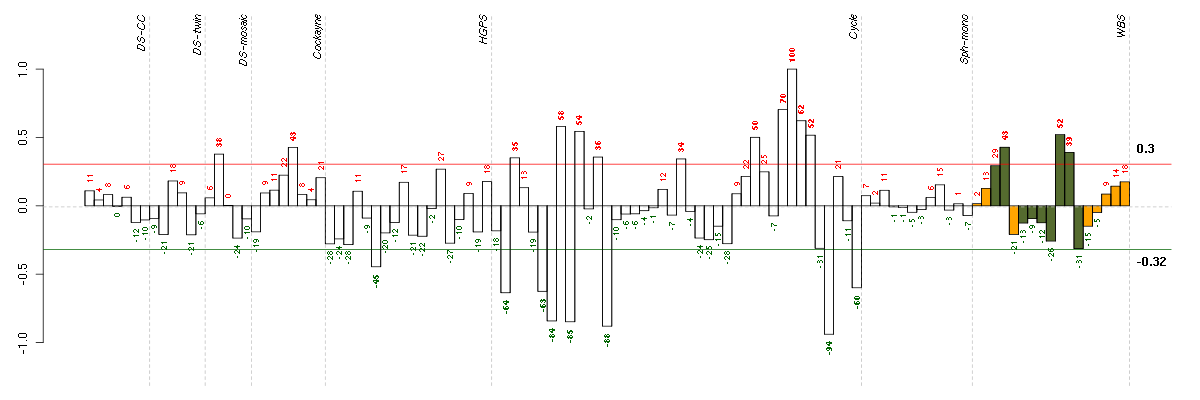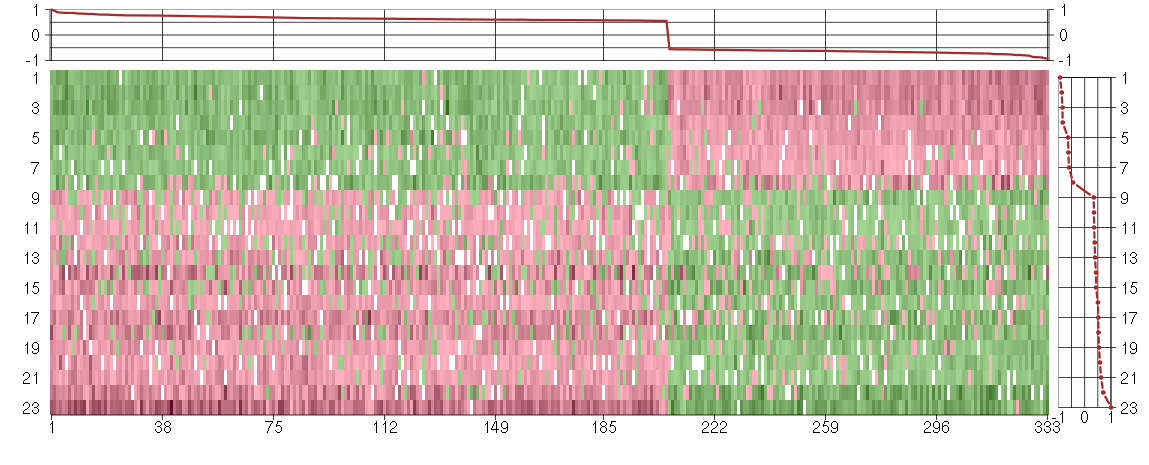

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
viral genome replication
Any process involved directly in viral genome replication, including viral nucleotide metabolism.
viral reproduction
The process by which a virus reproduces. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
RNA modification
The covalent alteration of one or more nucleotides within an RNA molecule to produce an RNA molecule with a sequence that differs from that coded genetically.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
base conversion or substitution editing
Any base modification or substitution events that result in alterations in the coding potential or structural properties of RNAs as a result of changes in the base-pairing properties of the modified ribonucleoside(s).
viral infectious cycle
A set of processes which all viruses follow to ensure survival; includes attachment and entry of the virus particle, decoding of genome information, translation of viral mRNA by host ribosomes, genome replication, and assembly and release of viral particles containing the genome.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
viral reproductive process
A reproductive process involved in viral reproduction. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological polymer, resulting in a change in its properties.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
regulation of viral genome replication
Any process that modulates the frequency, rate or extent of viral genome replication.
negative regulation of viral genome replication
Any process that stops, prevents or reduces the frequency, rate or extent of viral genome replication.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
negative regulation of viral reproduction
Any process that stops, prevents or reduces the frequency, rate or extent of the viral life cycle, the set of processes by which a virus reproduces and spreads among hosts.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of viral reproduction
Any process that modulates the rate or extent of the viral life cycle, the set of processes by which a virus reproduces and spreads among hosts.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
This term is the most general term possible
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
viral reproductive process
A reproductive process involved in viral reproduction. Usually, this is by infection of a host cell, replication of the viral genome, and assembly of progeny virus particles. In some cases the viral genetic material may integrate into the host genome and only subsequently, under particular circumstances, 'complete' its life cycle.
negative regulation of viral reproduction
Any process that stops, prevents or reduces the frequency, rate or extent of the viral life cycle, the set of processes by which a virus reproduces and spreads among hosts.
negative regulation of biological process
Any process that stops, prevents or reduces the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of viral reproduction
Any process that modulates the rate or extent of the viral life cycle, the set of processes by which a virus reproduces and spreads among hosts.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
The chemical reactions and pathways involving nucleobases, nucleosides, nucleotides and nucleic acids.
negative regulation of viral reproduction
Any process that stops, prevents or reduces the frequency, rate or extent of the viral life cycle, the set of processes by which a virus reproduces and spreads among hosts.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
regulation of viral genome replication
Any process that modulates the frequency, rate or extent of viral genome replication.
negative regulation of viral genome replication
Any process that stops, prevents or reduces the frequency, rate or extent of viral genome replication.
viral genome replication
Any process involved directly in viral genome replication, including viral nucleotide metabolism.
negative regulation of viral genome replication
Any process that stops, prevents or reduces the frequency, rate or extent of viral genome replication.
RNA metabolic process
The chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
RNA modification
The covalent alteration of one or more nucleotides within an RNA molecule to produce an RNA molecule with a sequence that differs from that coded genetically.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.66 ABHD3abhydrolase domain containing 3 (213017_at), score: -0.64 ABHD5abhydrolase domain containing 5 (218739_at), score: -0.65 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.61 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.7 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.72 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.71 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.76 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.75 ADORA2Badenosine A2b receptor (205891_at), score: 0.87 AGRNagrin (217419_x_at), score: 0.57 AHI1Abelson helper integration site 1 (221569_at), score: -0.69 AIM1absent in melanoma 1 (212543_at), score: 0.8 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.7 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.75 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.55 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.84 ANK2ankyrin 2, neuronal (202920_at), score: 0.6 ANXA10annexin A10 (210143_at), score: -0.78 AOX1aldehyde oxidase 1 (205083_at), score: 0.58 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.63 APOBEC3Gapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (204205_at), score: 0.64 APOL3apolipoprotein L, 3 (221087_s_at), score: 0.65 APOL6apolipoprotein L, 6 (219716_at), score: 0.75 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.62 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (218501_at), score: 0.64 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.68 ARMC9armadillo repeat containing 9 (219637_at), score: 0.66 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.76 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.59 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: -0.68 B9D1B9 protein domain 1 (210534_s_at), score: 0.74 BDKRB2bradykinin receptor B2 (205870_at), score: 0.72 BEX4brain expressed, X-linked 4 (215440_s_at), score: 0.58 BIN1bridging integrator 1 (210201_x_at), score: 0.66 BMP6bone morphogenetic protein 6 (206176_at), score: -0.62 BNC2basonuclin 2 (220272_at), score: 0.57 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: -0.68 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.61 BTG2BTG family, member 2 (201236_s_at), score: 0.76 C11orf41chromosome 11 open reading frame 41 (214772_at), score: -0.59 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.78 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.58 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.65 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.57 C4orf15chromosome 4 open reading frame 15 (210054_at), score: -0.61 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.58 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.61 CALB2calbindin 2 (205428_s_at), score: -0.76 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.8 CAMLGcalcium modulating ligand (203538_at), score: 0.6 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.6 CCNE2cyclin E2 (205034_at), score: -0.62 CCNG1cyclin G1 (208796_s_at), score: 0.67 CDC42EP2CDC42 effector protein (Rho GTPase binding) 2 (209850_s_at), score: 0.59 CDC6cell division cycle 6 homolog (S. cerevisiae) (203967_at), score: -0.58 CDCP1CUB domain containing protein 1 (218451_at), score: -0.57 CDH4cadherin 4, type 1, R-cadherin (retinal) (206866_at), score: -0.6 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.76 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.6 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.62 CLASP2cytoplasmic linker associated protein 2 (212306_at), score: -0.59 CLDN11claudin 11 (206908_s_at), score: 0.6 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.77 CPA4carboxypeptidase A4 (205832_at), score: 0.59 CPZcarboxypeptidase Z (211062_s_at), score: 0.59 CRBNcereblon (218142_s_at), score: 0.65 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.87 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: 0.76 DAAM2dishevelled associated activator of morphogenesis 2 (212793_at), score: 0.65 DAPK1death-associated protein kinase 1 (203139_at), score: 0.74 DCKdeoxycytidine kinase (203302_at), score: -0.71 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.72 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.65 DDX58DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (218943_s_at), score: 0.64 DDX60DEAD (Asp-Glu-Ala-Asp) box polypeptide 60 (218986_s_at), score: 0.67 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.81 DEPDC6DEP domain containing 6 (218858_at), score: 0.79 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.66 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.75 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.73 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.76 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.73 DPH5DPH5 homolog (S. cerevisiae) (219590_x_at), score: 0.59 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.59 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.72 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.62 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.66 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.67 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.61 EFNA5ephrin-A5 (214036_at), score: 0.64 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.67 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.62 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.72 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.6 EPHA4EPH receptor A4 (206114_at), score: -0.56 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.6 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.71 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.64 FBXL7F-box and leucine-rich repeat protein 7 (213249_at), score: 0.59 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.6 FICDFIC domain containing (219910_at), score: -0.62 FLT3LGfms-related tyrosine kinase 3 ligand (210607_at), score: 0.63 FNBP1formin binding protein 1 (212288_at), score: 0.77 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.69 FSTL3follistatin-like 3 (secreted glycoprotein) (203592_s_at), score: -0.58 FYNFYN oncogene related to SRC, FGR, YES (210105_s_at), score: 0.65 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.7 GALgalanin prepropeptide (214240_at), score: -0.65 GALNAC4S-6STB cell RAG associated protein (203066_at), score: 0.63 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.57 GDF15growth differentiation factor 15 (221577_x_at), score: 0.65 GDF5growth differentiation factor 5 (206614_at), score: 0.64 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.75 GKglycerol kinase (207387_s_at), score: -0.79 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.87 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: 0.71 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.72 GMPRguanosine monophosphate reductase (204187_at), score: 0.77 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.7 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.6 GRAMD1BGRAM domain containing 1B (212906_at), score: -0.64 GREM2gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) (220794_at), score: -0.6 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: -0.63 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.95 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.61 HERC6hect domain and RLD 6 (219352_at), score: 0.62 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.66 HHIPL2HHIP-like 2 (220283_at), score: -0.58 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.59 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.57 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.57 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.63 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.69 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.62 IFI35interferon-induced protein 35 (209417_s_at), score: 0.62 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.82 IFIT3interferon-induced protein with tetratricopeptide repeats 3 (204747_at), score: 0.59 IGBP1immunoglobulin (CD79A) binding protein 1 (202105_at), score: 0.6 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.67 IL33interleukin 33 (209821_at), score: -0.64 IRF1interferon regulatory factor 1 (202531_at), score: 0.87 IRF2interferon regulatory factor 2 (203275_at), score: 0.63 IVNS1ABPinfluenza virus NS1A binding protein (206245_s_at), score: -0.56 JUNDjun D proto-oncogene (203751_x_at), score: 0.57 JUPjunction plakoglobin (201015_s_at), score: 0.59 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.88 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.62 KIAA1305KIAA1305 (220911_s_at), score: 0.83 KIAA1462KIAA1462 (213316_at), score: 0.69 KLHDC2kelch domain containing 2 (217906_at), score: 0.58 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.71 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.62 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.73 LEPREL1leprecan-like 1 (218717_s_at), score: -0.61 LETMD1LETM1 domain containing 1 (207170_s_at), score: 0.59 LIMK2LIM domain kinase 2 (202193_at), score: -0.58 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.66 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.76 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.66 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.79 LOC399491LOC399491 protein (214035_x_at), score: 0.73 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.6 LPIN1lipin 1 (212274_at), score: 0.56 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.86 LRRC49leucine rich repeat containing 49 (219338_s_at), score: 0.57 LY75lymphocyte antigen 75 (205668_at), score: 0.65 LYRM1LYR motif containing 1 (203897_at), score: 0.58 LZTFL1leucine zipper transcription factor-like 1 (218437_s_at), score: 0.56 MAFv-maf musculoaponeurotic fibrosarcoma oncogene homolog (avian) (209348_s_at), score: 0.57 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 1 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.68 MAPRE2microtubule-associated protein, RP/EB family, member 2 (202501_at), score: 0.56 MBPmyelin basic protein (210136_at), score: 0.63 MC4Rmelanocortin 4 receptor (221467_at), score: -0.75 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.62 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.66 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.64 MKNK2MAP kinase interacting serine/threonine kinase 2 (218205_s_at), score: 0.61 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.8 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.8 MMP3matrix metallopeptidase 3 (stromelysin 1, progelatinase) (205828_at), score: -0.56 MRASmuscle RAS oncogene homolog (206538_at), score: -0.61 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.7 MSX2msh homeobox 2 (210319_x_at), score: 0.7 MT1Xmetallothionein 1X (204326_x_at), score: -0.6 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.59 MX1myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) (202086_at), score: 0.66 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.77 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.7 NCALDneurocalcin delta (211685_s_at), score: -0.75 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.61 NEFMneurofilament, medium polypeptide (205113_at), score: -0.91 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.69 NINJ1ninjurin 1 (203045_at), score: 0.62 NMIN-myc (and STAT) interactor (203964_at), score: 0.74 NOX4NADPH oxidase 4 (219773_at), score: -0.59 NPnucleoside phosphorylase (201695_s_at), score: -0.59 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.57 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.8 NPR3natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) (219789_at), score: -0.58 NPTX1neuronal pentraxin I (204684_at), score: -0.73 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.69 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.67 NTMneurotrimin (222020_s_at), score: -0.76 NUP160nucleoporin 160kDa (212709_at), score: -0.65 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: 0.58 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.85 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.61 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.79 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.8 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.68 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.69 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.71 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.59 PARP12poly (ADP-ribose) polymerase family, member 12 (218543_s_at), score: 0.6 PCDH9protocadherin 9 (219737_s_at), score: -0.87 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.66 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.65 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.77 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.59 PDGFRBplatelet-derived growth factor receptor, beta polypeptide (202273_at), score: 0.61 PHC1polyhomeotic homolog 1 (Drosophila) (218338_at), score: 0.63 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.61 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.66 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.59 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.55 PLEKHA5pleckstrin homology domain containing, family A member 5 (220952_s_at), score: 0.59 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.64 PNRC1proline-rich nuclear receptor coactivator 1 (209034_at), score: 0.66 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.77 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.72 PPLperiplakin (203407_at), score: 0.71 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: -0.63 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.62 PRR5proline rich 5 (renal) (47069_at), score: 0.59 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.63 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.66 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.57 PTGER4prostaglandin E receptor 4 (subtype EP4) (204897_at), score: 0.61 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.67 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.57 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.57 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.57 PYGLphosphorylase, glycogen, liver (202990_at), score: 0.59 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.65 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.61 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.7 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.58 RCAN2regulator of calcineurin 2 (203498_at), score: 0.72 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.71 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: 0.62 RNF220ring finger protein 220 (219988_s_at), score: 0.58 RNF44ring finger protein 44 (203286_at), score: 0.64 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: 0.64 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.74 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.78 RPL13Aribosomal protein L13a (200715_x_at), score: 0.72 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.59 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.67 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.71 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.63 SCG5secretogranin V (7B2 protein) (203889_at), score: -0.62 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.68 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.57 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.82 SELENBP1selenium binding protein 1 (214433_s_at), score: 0.77 SEPP1selenoprotein P, plasma, 1 (201427_s_at), score: 0.57 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.65 SESN1sestrin 1 (218346_s_at), score: 0.73 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.6 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.64 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.58 SIX2SIX homeobox 2 (206510_at), score: 0.68 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.72 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.63 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.68 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.62 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.62 SMAD3SMAD family member 3 (218284_at), score: 0.83 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.56 SNNstannin (218032_at), score: 0.62 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.56 SPATA20spermatogenesis associated 20 (218164_at), score: 0.63 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.62 SSTR1somatostatin receptor 1 (208482_at), score: -0.68 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.77 STIM1stromal interaction molecule 1 (202764_at), score: 0.6 SVILsupervillin (202565_s_at), score: 0.59 SYNJ1synaptojanin 1 (212990_at), score: -0.67 SYNPOsynaptopodin (202796_at), score: 0.71 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.73 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.87 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.84 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.67 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.63 TLR3toll-like receptor 3 (206271_at), score: 0.88 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.61 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.65 TMEM140transmembrane protein 140 (218999_at), score: 0.61 TMEM2transmembrane protein 2 (218113_at), score: -0.68 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.71 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.73 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.68 TNS3tensin 3 (217853_at), score: 0.78 TP53tumor protein p53 (201746_at), score: 0.89 TRIM21tripartite motif-containing 21 (204804_at), score: 0.76 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.62 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.64 TSKUtsukushin (218245_at), score: 0.61 TSPAN13tetraspanin 13 (217979_at), score: -0.82 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.62 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.75 UBXN1UBX domain protein 1 (201871_s_at), score: 0.65 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.69 ULBP2UL16 binding protein 2 (221291_at), score: -0.92 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.64 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.73 WBP2WW domain binding protein 2 (209117_at), score: 0.64 WDR4WD repeat domain 4 (221632_s_at), score: -0.65 WDR42AWD repeat domain 42A (202249_s_at), score: 0.58 WDR6WD repeat domain 6 (217734_s_at), score: 0.64 XPCxeroderma pigmentosum, complementation group C (209375_at), score: 0.56 XYLT1xylosyltransferase I (213725_x_at), score: -0.73 YDD19YDD19 protein (37079_at), score: -0.56 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.61 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.6 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.61 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.57 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.74 ZNF358zinc finger protein 358 (219379_x_at), score: 0.55 ZNF580zinc finger protein 580 (220748_s_at), score: 0.6 ZNF706zinc finger protein 706 (218059_at), score: 0.55
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690256.cel | 6 | 5 | HGPS | hgu133a | none | GMO8398C |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46B.CEL | 2 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 2 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-GEOD-3407-raw-cel-1437949721.cel | 5 | 4 | Cockayne | hgu133a | CS | CSB |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486331.cel | 35 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485791.cel | 8 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486311.cel | 34 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |