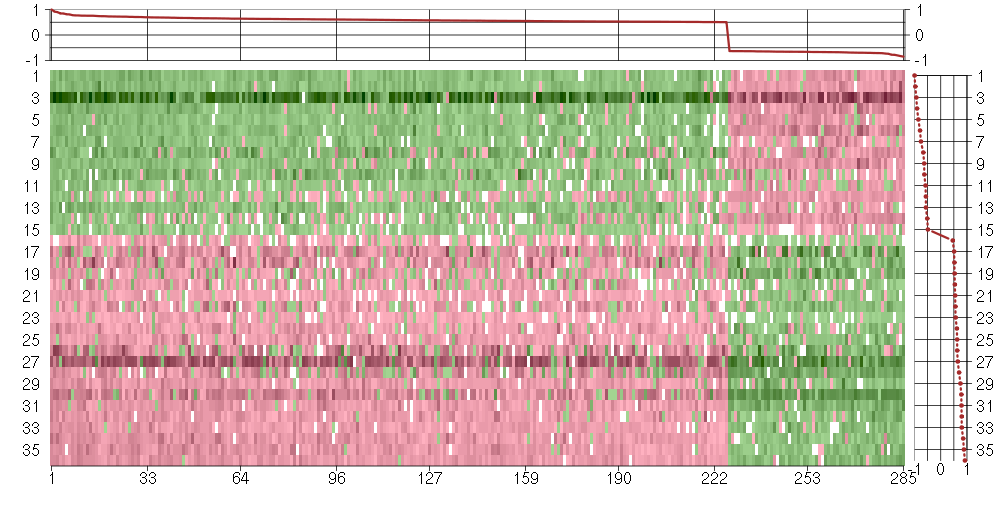

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
receptor metabolic process
The chemical reactions and pathways involving a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
all
This term is the most general term possible


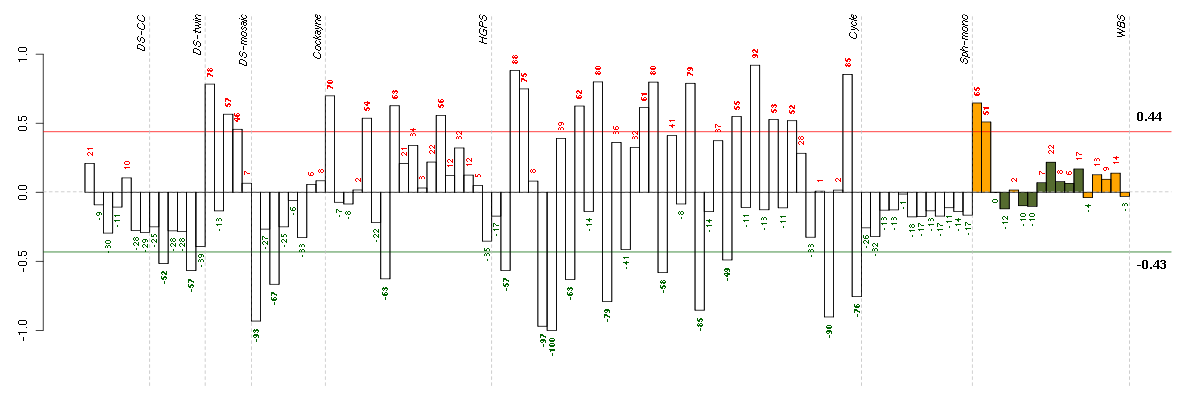
ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.76 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.67 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.57 AESamino-terminal enhancer of split (217729_s_at), score: 0.64 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.68 AGRNagrin (217419_x_at), score: 0.63 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.53 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.73 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.59 APBB1amyloid beta (A4) precursor protein-binding, family B, member 1 (Fe65) (202652_at), score: 0.53 ARHGDIARho GDP dissociation inhibitor (GDI) alpha (213606_s_at), score: 0.54 ARSAarylsulfatase A (204443_at), score: 0.84 ATG9AATG9 autophagy related 9 homolog A (S. cerevisiae) (202492_at), score: 0.55 ATN1atrophin 1 (40489_at), score: 0.62 ATP13A2ATPase type 13A2 (218608_at), score: 0.5 ATP6AP1ATPase, H+ transporting, lysosomal accessory protein 1 (207809_s_at), score: 0.51 B3GAT3beta-1,3-glucuronyltransferase 3 (glucuronosyltransferase I) (203452_at), score: 0.56 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.59 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.76 BAT2LHLA-B associated transcript 2-like (212068_s_at), score: 0.6 BCL2L1BCL2-like 1 (215037_s_at), score: 0.61 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.72 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.7 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.54 C19orf22chromosome 19 open reading frame 22 (221764_at), score: 0.6 C20orf149chromosome 20 open reading frame 149 (218010_x_at), score: 0.57 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.7 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.67 C9orf16chromosome 9 open reading frame 16 (204480_s_at), score: 0.51 CABIN1calcineurin binding protein 1 (37652_at), score: 0.56 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.75 CAPN1calpain 1, (mu/I) large subunit (200752_s_at), score: 0.58 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.71 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.78 CDC42EP1CDC42 effector protein (Rho GTPase binding) 1 (204693_at), score: 0.7 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.51 CHMP5chromatin modifying protein 5 (218085_at), score: -0.66 CHPFchondroitin polymerizing factor (202175_at), score: 0.64 CHST3carbohydrate (chondroitin 6) sulfotransferase 3 (209834_at), score: 0.51 CICcapicua homolog (Drosophila) (212784_at), score: 0.69 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.63 CLIP2CAP-GLY domain containing linker protein 2 (211031_s_at), score: 0.55 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.62 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.66 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.57 CORO1Bcoronin, actin binding protein, 1B (64486_at), score: 0.58 CRCPCGRP receptor component (203898_at), score: -0.63 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.58 CST3cystatin C (201360_at), score: 0.51 CUL7cullin 7 (36084_at), score: 0.61 DBN1drebrin 1 (217025_s_at), score: 0.53 DEAF1deformed epidermal autoregulatory factor 1 (Drosophila) (209407_s_at), score: 0.6 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: -0.71 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.63 DNAJC4DnaJ (Hsp40) homolog, subfamily C, member 4 (206782_s_at), score: 0.57 DNM2dynamin 2 (202253_s_at), score: 0.63 DOK4docking protein 4 (209691_s_at), score: 0.61 DOPEY1dopey family member 1 (40612_at), score: -0.8 DRG2developmentally regulated GTP binding protein 2 (203267_s_at), score: 0.59 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.65 DUS2Ldihydrouridine synthase 2-like, SMM1 homolog (S. cerevisiae) (47105_at), score: -0.65 DUSP3dual specificity phosphatase 3 (201537_s_at), score: 0.52 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.67 EFHD2EF-hand domain family, member D2 (217992_s_at), score: 0.53 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.66 EHD2EH-domain containing 2 (221870_at), score: 0.56 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.7 EVLEnah/Vasp-like (217838_s_at), score: 0.53 F8A1coagulation factor VIII-associated (intronic transcript) 1 (203274_at), score: 0.53 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.52 FAM115Afamily with sequence similarity 115, member A (212981_s_at), score: -0.63 FAM176Bfamily with sequence similarity 176, member B (220134_x_at), score: 0.61 FAM65Afamily with sequence similarity 65, member A (45749_at), score: 0.63 FBXO17F-box protein 17 (220233_at), score: 0.58 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.64 FCF1FCF1 small subunit (SSU) processome component homolog (S. cerevisiae) (219927_at), score: -0.69 FETUBfetuin B (214417_s_at), score: -0.63 FHOD1formin homology 2 domain containing 1 (218530_at), score: 0.61 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.74 FKSG2apoptosis inhibitor (208588_at), score: -0.67 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.62 FOXK2forkhead box K2 (203064_s_at), score: 0.7 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: 0.65 GNAI2guanine nucleotide binding protein (G protein), alpha inhibiting activity polypeptide 2 (201040_at), score: 0.53 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.51 GRINAglutamate receptor, ionotropic, N-methyl D-aspartate-associated protein 1 (glutamate binding) (212090_at), score: 0.72 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.63 GTPBP1GTP binding protein 1 (219357_at), score: 0.54 HAB1B1 for mucin (215778_x_at), score: -0.65 HDAC5histone deacetylase 5 (202455_at), score: 0.6 HLA-Emajor histocompatibility complex, class I, E (200904_at), score: 0.63 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.71 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.64 HPS1Hermansky-Pudlak syndrome 1 (217354_s_at), score: 0.52 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.68 HSF1heat shock transcription factor 1 (202344_at), score: 0.68 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.76 IGFBP4insulin-like growth factor binding protein 4 (201508_at), score: 0.54 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.66 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.65 INF2inverted formin, FH2 and WH2 domain containing (218144_s_at), score: 0.52 INTS1integrator complex subunit 1 (212212_s_at), score: 0.53 ITGA3integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) (201474_s_at), score: 0.53 JUNDjun D proto-oncogene (203751_x_at), score: 0.81 JUPjunction plakoglobin (201015_s_at), score: 0.76 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.69 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.54 KCTD3potassium channel tetramerisation domain containing 3 (217894_at), score: -0.63 KDELR1KDEL (Lys-Asp-Glu-Leu) endoplasmic reticulum protein retention receptor 1 (200922_at), score: 0.6 KIAA1305KIAA1305 (220911_s_at), score: 0.64 KIAA1539KIAA1539 (207765_s_at), score: 0.55 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.67 KLHL26kelch-like 26 (Drosophila) (219354_at), score: 0.61 LEPREL2leprecan-like 2 (204854_at), score: 0.64 LIMS2LIM and senescent cell antigen-like domains 2 (220765_s_at), score: 0.61 LMF2lipase maturation factor 2 (212682_s_at), score: 0.59 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.63 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.71 LPPR2lipid phosphate phosphatase-related protein type 2 (218509_at), score: 0.55 LRFN4leucine rich repeat and fibronectin type III domain containing 4 (219491_at), score: 0.5 LRP5low density lipoprotein receptor-related protein 5 (209468_at), score: 0.53 LRRC40leucine rich repeat containing 40 (218577_at), score: -0.67 LRRC41leucine rich repeat containing 41 (201932_at), score: 0.54 LTBP3latent transforming growth factor beta binding protein 3 (219922_s_at), score: 0.66 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.78 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.9 MAP2K2mitogen-activated protein kinase kinase 2 (213490_s_at), score: 0.68 MAP3K11mitogen-activated protein kinase kinase kinase 11 (203652_at), score: 0.58 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.65 MAP4microtubule-associated protein 4 (200836_s_at), score: 0.61 MAP7D1MAP7 domain containing 1 (217943_s_at), score: 0.67 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.62 MBOAT7membrane bound O-acyltransferase domain containing 7 (209179_s_at), score: 0.55 MED15mediator complex subunit 15 (222175_s_at), score: 0.57 MFSD10major facilitator superfamily domain containing 10 (209215_at), score: 0.54 MGAT1mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase (201126_s_at), score: 0.66 MGAT4Bmannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme B (220189_s_at), score: 0.64 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.56 MKL1megakaryoblastic leukemia (translocation) 1 (212748_at), score: 0.56 MRPS2mitochondrial ribosomal protein S2 (218001_at), score: 0.54 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.69 MUL1mitochondrial E3 ubiquitin ligase 1 (218246_at), score: 0.54 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.68 MYO9Bmyosin IXB (217297_s_at), score: 0.58 NACAP1nascent-polypeptide-associated complex alpha polypeptide pseudogene 1 (211445_x_at), score: -0.69 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.66 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.52 NCDNneurochondrin (209556_at), score: 0.6 NCKAP1NCK-associated protein 1 (217465_at), score: -0.83 NCOR2nuclear receptor co-repressor 2 (207760_s_at), score: 0.6 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.65 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.56 NINJ1ninjurin 1 (203045_at), score: 0.52 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: 0.58 NOVA2neuro-oncological ventral antigen 2 (206477_s_at), score: -0.67 NPDC1neural proliferation, differentiation and control, 1 (218086_at), score: 0.65 NUCB1nucleobindin 1 (200646_s_at), score: 0.52 OBSL1obscurin-like 1 (212775_at), score: 0.52 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (201282_at), score: 0.61 PARVBparvin, beta (204629_at), score: 0.71 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.77 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.72 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.67 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.64 PCDHGC3protocadherin gamma subfamily C, 3 (211066_x_at), score: 0.52 PDE7Bphosphodiesterase 7B (220343_at), score: -0.65 PDLIM7PDZ and LIM domain 7 (enigma) (203370_s_at), score: 0.5 PHF7PHD finger protein 7 (215622_x_at), score: -0.68 PHLDA3pleckstrin homology-like domain, family A, member 3 (218634_at), score: 0.54 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.55 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.57 PIK3R2phosphoinositide-3-kinase, regulatory subunit 2 (beta) (207105_s_at), score: 0.63 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.59 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.92 PKD1polycystic kidney disease 1 (autosomal dominant) (202328_s_at), score: 0.53 PKN1protein kinase N1 (202161_at), score: 0.72 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.56 PLCD1phospholipase C, delta 1 (205125_at), score: 0.51 PLD3phospholipase D family, member 3 (201050_at), score: 0.7 PLEC1plectin 1, intermediate filament binding protein 500kDa (216971_s_at), score: 0.53 PLK3polo-like kinase 3 (Drosophila) (204958_at), score: 0.54 PLSCR3phospholipid scramblase 3 (218828_at), score: 0.6 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.72 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.61 POLRMTpolymerase (RNA) mitochondrial (DNA directed) (203782_s_at), score: 0.53 POM121POM121 membrane glycoprotein (rat) (212178_s_at), score: 0.59 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.53 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.57 PRKACAprotein kinase, cAMP-dependent, catalytic, alpha (202801_at), score: 0.56 PRKCDprotein kinase C, delta (202545_at), score: 0.66 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: 0.57 PTPN14protein tyrosine phosphatase, non-receptor type 14 (205503_at), score: 0.53 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.59 PXNpaxillin (211823_s_at), score: 0.77 PYGBphosphorylase, glycogen; brain (201481_s_at), score: 0.56 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.67 RAB11FIP3RAB11 family interacting protein 3 (class II) (203933_at), score: 0.52 RANBP3RAN binding protein 3 (210120_s_at), score: 0.63 RBM15BRNA binding motif protein 15B (202689_at), score: 0.64 RELAv-rel reticuloendotheliosis viral oncogene homolog A (avian) (209878_s_at), score: 0.62 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.51 RFNGRFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (212968_at), score: 0.52 RHOGras homolog gene family, member G (rho G) (203175_at), score: 0.52 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.65 RNF126ring finger protein 126 (205748_s_at), score: 0.52 RNF130ring finger protein 130 (217865_at), score: 0.56 RNF40ring finger protein 40 (206845_s_at), score: 0.52 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.71 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.67 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.74 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.85 RPS25ribosomal protein S25 (200091_s_at), score: -0.66 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.55 SBF1SET binding factor 1 (39835_at), score: 0.64 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.68 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.85 SCAPSREBF chaperone (212329_at), score: 0.53 SDAD1SDA1 domain containing 1 (218607_s_at), score: 0.56 SDC3syndecan 3 (202898_at), score: 0.66 SFRP1secreted frizzled-related protein 1 (202036_s_at), score: 0.53 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.71 SHBSrc homology 2 domain containing adaptor protein B (204657_s_at), score: 0.62 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.63 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.63 SIN3BSIN3 homolog B, transcription regulator (yeast) (209352_s_at), score: 0.53 SIP1survival of motor neuron protein interacting protein 1 (205063_at), score: -0.66 SLC25A1solute carrier family 25 (mitochondrial carrier; citrate transporter), member 1 (210010_s_at), score: 0.57 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (218985_at), score: 0.51 SLC38A10solute carrier family 38, member 10 (212890_at), score: 0.52 SLC9A1solute carrier family 9 (sodium/hydrogen exchanger), member 1 (209453_at), score: 0.58 SLC9A3R2solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 (209830_s_at), score: 0.67 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.73 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.63 SMTNsmoothelin (209427_at), score: 0.51 SMYD5SMYD family member 5 (209516_at), score: 0.53 SNAPC2small nuclear RNA activating complex, polypeptide 2, 45kDa (204104_at), score: 0.51 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.69 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.58 SP3Sp3 transcription factor (213168_at), score: -0.64 SPATA20spermatogenesis associated 20 (218164_at), score: 0.65 SPG7spastic paraplegia 7 (pure and complicated autosomal recessive) (202104_s_at), score: 0.51 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: 0.57 SSBP3single stranded DNA binding protein 3 (217991_x_at), score: 0.74 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.59 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.66 SUPT6Hsuppressor of Ty 6 homolog (S. cerevisiae) (208831_x_at), score: 0.6 SYNPOsynaptopodin (202796_at), score: 0.51 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.65 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.75 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.54 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.55 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.56 TGFB1transforming growth factor, beta 1 (203085_s_at), score: 0.55 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.7 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.6 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.67 TMEM184Ctransmembrane protein 184C (219074_at), score: -0.64 TMEM222transmembrane protein 222 (221512_at), score: 0.52 TMEM8transmembrane protein 8 (five membrane-spanning domains) (221882_s_at), score: 0.57 TMSB4Ythymosin beta 4, Y-linked (206769_at), score: -0.69 TNFRSF8tumor necrosis factor receptor superfamily, member 8 (206729_at), score: -0.67 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.73 TSKUtsukushin (218245_at), score: 0.74 TTC15tetratricopeptide repeat domain 15 (203122_at), score: 0.55 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.77 TXLNAtaxilin alpha (212300_at), score: 0.61 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.67 UBTD1ubiquitin domain containing 1 (219172_at), score: 0.69 UBXN6UBX domain protein 6 (220757_s_at), score: 0.62 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.52 UPF2UPF2 regulator of nonsense transcripts homolog (yeast) (203519_s_at), score: -0.66 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.71 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.65 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (219810_at), score: -0.66 VEGFBvascular endothelial growth factor B (203683_s_at), score: 1 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: -0.64 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.81 WBP2WW domain binding protein 2 (209117_at), score: 0.69 WDR42AWD repeat domain 42A (202249_s_at), score: 0.62 XAB2XPA binding protein 2 (218110_at), score: 0.52 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.61 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.58 ZNF282zinc finger protein 282 (212892_at), score: 0.65 ZNF580zinc finger protein 580 (220748_s_at), score: 0.62 ZYXzyxin (200808_s_at), score: 0.56
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949557.cel | 1 | 4 | Cockayne | hgu133a | CS | eGFP |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3407-raw-cel-1437949655.cel | 3 | 4 | Cockayne | hgu133a | none | CSB |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690272.cel | 7 | 5 | HGPS | hgu133a | HGPS | AG11498 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 5CTwin.CEL | 5 | 2 | DS-twin | hgu133plus2 | Down | DS-twin 5 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 2Twin.CEL | 2 | 2 | DS-twin | hgu133plus2 | none | DS-twin 2 |
| E-TABM-263-raw-cel-1515486151.cel | 26 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 47B.CEL | 4 | 3 | DS-mosaic | hgu133plus2 | Down mosaic | DS-mosaic 4 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690248.cel | 5 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690376.cel | 13 | 5 | HGPS | hgu133a | HGPS | AG11513 |
| 46C.CEL | 3 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 3 |
| E-TABM-263-raw-cel-1515485971.cel | 17 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-GEOD-3860-raw-cel-1561690304.cel | 8 | 5 | HGPS | hgu133a | none | GMO8398C |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-GEOD-3860-raw-cel-1561690199.cel | 1 | 5 | HGPS | hgu133a | none | GM0316B |
| E-TABM-263-raw-cel-1515485711.cel | 4 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 46A.CEL | 1 | 3 | DS-mosaic | hgu133plus2 | none | DS-mosaic 1 |
| E-TABM-263-raw-cel-1515486071.cel | 22 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485991.cel | 18 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486411.cel | 39 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |