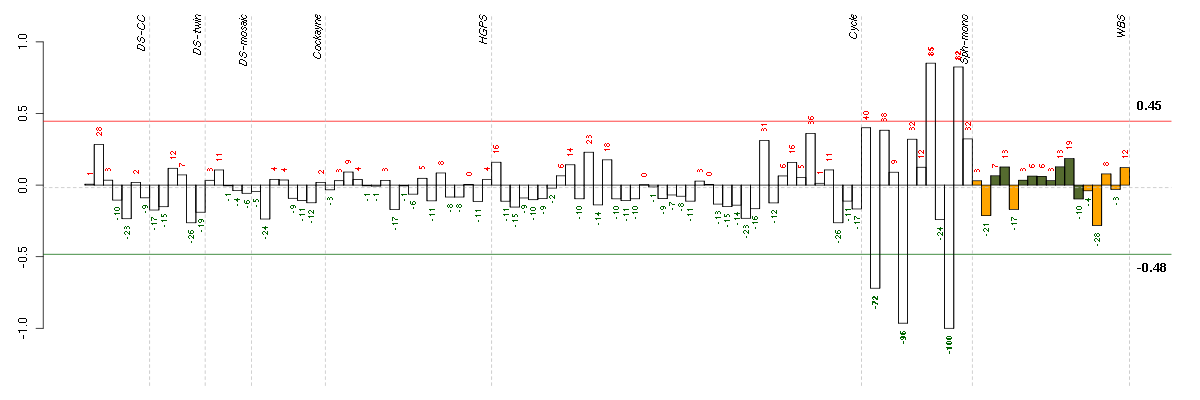

Under-expression is coded with green,
over-expression with red color.



ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: 0.75 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (204567_s_at), score: 0.76 ABI1abl-interactor 1 (209028_s_at), score: 0.66 ACACAacetyl-Coenzyme A carboxylase alpha (212186_at), score: -0.81 ACAT1acetyl-Coenzyme A acetyltransferase 1 (205412_at), score: -0.62 ACTBactin, beta (AFFX-HSAC07/X00351_5_at), score: -0.7 ACVR1activin A receptor, type I (203935_at), score: 0.54 ADFPadipose differentiation-related protein (209122_at), score: 0.59 ADNPactivity-dependent neuroprotector homeobox (201773_at), score: 0.58 ADO2-aminoethanethiol (cysteamine) dioxygenase (212500_at), score: 0.53 ADORA2Aadenosine A2a receptor (205013_s_at), score: 0.64 ADORA2Badenosine A2b receptor (205891_at), score: -0.65 AHI1Abelson helper integration site 1 (221569_at), score: 0.6 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.62 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: -0.79 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: -0.65 AMPD3adenosine monophosphate deaminase (isoform E) (207992_s_at), score: 0.68 ANKHankylosis, progressive homolog (mouse) (220076_at), score: 0.71 ANKRD10ankyrin repeat domain 10 (218093_s_at), score: 0.73 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: -0.63 APH1Aanterior pharynx defective 1 homolog A (C. elegans) (218389_s_at), score: -0.63 APODapolipoprotein D (201525_at), score: 0.56 APOL2apolipoprotein L, 2 (221653_x_at), score: -0.7 AREGamphiregulin (205239_at), score: 0.53 ARHGAP12Rho GTPase activating protein 12 (207606_s_at), score: 0.7 ARHGAP24Rho GTPase activating protein 24 (221030_s_at), score: 0.63 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: 0.55 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: 0.61 ARL8BADP-ribosylation factor-like 8B (217852_s_at), score: 0.58 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: 0.65 ARMETarginine-rich, mutated in early stage tumors (202655_at), score: -0.62 ASB9ankyrin repeat and SOCS box-containing 9 (205673_s_at), score: -0.81 ATF1activating transcription factor 1 (222103_at), score: 0.57 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: -0.8 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: 0.61 ATP5G1ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C1 (subunit 9) (208972_s_at), score: -0.64 ATP5G3ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C3 (subunit 9) (207507_s_at), score: -0.62 AVL9AVL9 homolog (S. cerevisiase) (212474_at), score: 0.54 BACH1BTB and CNC homology 1, basic leucine zipper transcription factor 1 (204194_at), score: 0.61 BACH2BTB and CNC homology 1, basic leucine zipper transcription factor 2 (221234_s_at), score: 0.57 BANF1barrier to autointegration factor 1 (210125_s_at), score: -0.65 BCL6B-cell CLL/lymphoma 6 (203140_at), score: 0.55 BDKRB2bradykinin receptor B2 (205870_at), score: 0.53 BEX4brain expressed, X-linked 4 (215440_s_at), score: 0.61 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: 0.59 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: -0.72 BMPR1Abone morphogenetic protein receptor, type IA (204832_s_at), score: 0.58 BTAF1BTAF1 RNA polymerase II, B-TFIID transcription factor-associated, 170kDa (Mot1 homolog, S. cerevisiae) (209430_at), score: 0.63 C10orf97chromosome 10 open reading frame 97 (218297_at), score: 0.52 C14orf138chromosome 14 open reading frame 138 (218940_at), score: 0.54 C14orf147chromosome 14 open reading frame 147 (212460_at), score: 0.58 C14orf169chromosome 14 open reading frame 169 (219526_at), score: 0.67 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: -0.64 C1orf54chromosome 1 open reading frame 54 (219506_at), score: 0.55 C1QBPcomplement component 1, q subcomponent binding protein (208910_s_at), score: -0.7 C20orf39chromosome 20 open reading frame 39 (219310_at), score: -0.67 C4orf16chromosome 4 open reading frame 16 (219023_at), score: 0.57 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: 0.66 C6orf47chromosome 6 open reading frame 47 (204968_at), score: -0.64 C7orf25chromosome 7 open reading frame 25 (53202_at), score: -0.61 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: 0.73 C9orf78chromosome 9 open reading frame 78 (218116_at), score: 0.6 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: 0.53 CACYBPcalcyclin binding protein (210691_s_at), score: -0.64 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: 0.6 CBR3carbonyl reductase 3 (205379_at), score: -0.64 CCDC44coiled-coil domain containing 44 (221069_s_at), score: -0.63 CCL8chemokine (C-C motif) ligand 8 (214038_at), score: 0.57 CCNFcyclin F (204826_at), score: -0.69 CCNT2cyclin T2 (204645_at), score: 0.69 CCT5chaperonin containing TCP1, subunit 5 (epsilon) (208696_at), score: -0.61 CCT7chaperonin containing TCP1, subunit 7 (eta) (200812_at), score: -0.66 CD24CD24 molecule (209771_x_at), score: -0.72 CDC37L1cell division cycle 37 homolog (S. cerevisiae)-like 1 (219343_at), score: 0.52 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.61 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: -0.73 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: -0.67 CENPIcentromere protein I (214804_at), score: -0.63 CENPNcentromere protein N (219555_s_at), score: -0.59 CEP170centrosomal protein 170kDa (212746_s_at), score: 0.58 CGRRF1cell growth regulator with ring finger domain 1 (204605_at), score: 0.53 CHCHD3coiled-coil-helix-coiled-coil-helix domain containing 3 (217972_at), score: -0.64 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.64 CHMP1Achromatin modifying protein 1A (201933_at), score: -0.6 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.67 CHST6carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 (221059_s_at), score: -0.64 CLDND1claudin domain containing 1 (208925_at), score: 0.52 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.57 CLIC1chloride intracellular channel 1 (208659_at), score: -0.66 CLK1CDC-like kinase 1 (214683_s_at), score: 0.61 CNIH3cornichon homolog 3 (Drosophila) (214841_at), score: 0.59 CNOT1CCR4-NOT transcription complex, subunit 1 (200860_s_at), score: -0.61 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.53 COL3A1collagen, type III, alpha 1 (215076_s_at), score: 0.64 CORO2Acoronin, actin binding protein, 2A (205538_at), score: -0.65 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: 0.67 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: 0.86 CSNK1G3casein kinase 1, gamma 3 (220768_s_at), score: 0.52 CTPSCTP synthase (202613_at), score: -0.68 CTSHcathepsin H (202295_s_at), score: -0.63 CTSKcathepsin K (202450_s_at), score: 0.64 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: 0.56 CXCL1chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) (204470_at), score: 0.64 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: 0.61 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.52 CXCL5chemokine (C-X-C motif) ligand 5 (214974_x_at), score: 0.57 CYB561cytochrome b-561 (209163_at), score: -0.63 DACT1dapper, antagonist of beta-catenin, homolog 1 (Xenopus laevis) (219179_at), score: 0.54 DCBLD2discoidin, CUB and LCCL domain containing 2 (213865_at), score: -0.62 DCNdecorin (211896_s_at), score: 0.6 DDTD-dopachrome tautomerase (202929_s_at), score: -0.59 DFNA5deafness, autosomal dominant 5 (203695_s_at), score: -0.59 DHCR2424-dehydrocholesterol reductase (200862_at), score: -0.65 DLEU1deleted in lymphocytic leukemia 1 (non-protein coding) (205677_s_at), score: -0.59 DNAJA1DnaJ (Hsp40) homolog, subfamily A, member 1 (200880_at), score: 0.52 DNAJA3DnaJ (Hsp40) homolog, subfamily A, member 3 (205963_s_at), score: -0.62 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: -0.64 DOCK4dedicator of cytokinesis 4 (205003_at), score: 0.54 DPYSL3dihydropyrimidinase-like 3 (201430_s_at), score: -0.62 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: -0.59 DVL3dishevelled, dsh homolog 3 (Drosophila) (201908_at), score: 0.53 E2F1E2F transcription factor 1 (204947_at), score: -0.67 EAPPE2F-associated phosphoprotein (202623_at), score: 0.59 EBNA1BP2EBNA1 binding protein 2 (201323_at), score: -0.65 EDNRAendothelin receptor type A (204464_s_at), score: 0.69 EGR2early growth response 2 (Krox-20 homolog, Drosophila) (205249_at), score: 0.52 EIF1Beukaryotic translation initiation factor 1B (201738_at), score: 0.58 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: -0.79 EIF4A3eukaryotic translation initiation factor 4A, isoform 3 (201303_at), score: -0.62 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: 0.52 EIF5A2eukaryotic translation initiation factor 5A2 (220198_s_at), score: 0.6 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: 0.57 EMDemerin (209477_at), score: -0.65 EMG1EMG1 nucleolar protein homolog (S. cerevisiae) (209233_at), score: -0.6 EMP1epithelial membrane protein 1 (201325_s_at), score: 0.74 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: 0.61 EPAS1endothelial PAS domain protein 1 (200878_at), score: 0.57 EPHA2EPH receptor A2 (203499_at), score: -0.63 ERBB2IPerbb2 interacting protein (217941_s_at), score: 0.55 EREGepiregulin (205767_at), score: 0.58 EVI5ecotropic viral integration site 5 (209717_at), score: 0.54 EXO1exonuclease 1 (204603_at), score: -0.61 FADS1fatty acid desaturase 1 (208962_s_at), score: -0.7 FADS2fatty acid desaturase 2 (202218_s_at), score: -0.6 FAM8A1family with sequence similarity 8, member A1 (203420_at), score: 0.52 FANCAFanconi anemia, complementation group A (203805_s_at), score: -0.59 FBXO3F-box protein 3 (218432_at), score: 0.62 FDFT1farnesyl-diphosphate farnesyltransferase 1 (210950_s_at), score: -0.62 FDPSfarnesyl diphosphate synthase (farnesyl pyrophosphate synthetase, dimethylallyltranstransferase, geranyltranstransferase) (201275_at), score: -0.65 FJX1four jointed box 1 (Drosophila) (219522_at), score: 0.56 FKBP4FK506 binding protein 4, 59kDa (200895_s_at), score: -0.59 FLJ10038hypothetical protein FLJ10038 (205510_s_at), score: 0.71 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: 0.67 FNDC3Afibronectin type III domain containing 3A (202304_at), score: 0.53 FOXN3forkhead box N3 (218031_s_at), score: 0.68 FOXO1forkhead box O1 (202724_s_at), score: 0.7 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209892_at), score: 0.6 GABARAPL2GABA(A) receptor-associated protein-like 2 (209046_s_at), score: 0.71 GALCgalactosylceramidase (204417_at), score: 0.53 GALNSgalactosamine (N-acetyl)-6-sulfate sulfatase (206335_at), score: -0.59 GAPDHglyceraldehyde-3-phosphate dehydrogenase (AFFX-HUMGAPDH/M33197_M_at), score: -0.61 GARNL1GTPase activating Rap/RanGAP domain-like 1 (214855_s_at), score: 0.71 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: 0.72 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.52 GEMIN6gem (nuclear organelle) associated protein 6 (219539_at), score: -0.59 GLAgalactosidase, alpha (214430_at), score: -0.59 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: 0.7 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: 0.51 GOLGA7golgi autoantigen, golgin subfamily a, 7 (217819_at), score: 0.53 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: 0.73 GPM6Bglycoprotein M6B (209170_s_at), score: 0.62 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.58 GPR137BG protein-coupled receptor 137B (204137_at), score: 0.63 GPR177G protein-coupled receptor 177 (221958_s_at), score: 0.56 GPR89BG protein-coupled receptor 89B (220642_x_at), score: -0.62 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: -0.59 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: 1 GRK5G protein-coupled receptor kinase 5 (204396_s_at), score: 0.57 GRWD1glutamate-rich WD repeat containing 1 (221549_at), score: -0.67 GSTP1glutathione S-transferase pi 1 (200824_at), score: -0.65 GYG2glycogenin 2 (215695_s_at), score: -0.59 H2AFJH2A histone family, member J (220936_s_at), score: 0.55 HBP1HMG-box transcription factor 1 (209102_s_at), score: 0.59 HBXIPhepatitis B virus x interacting protein (202299_s_at), score: 0.52 HECAheadcase homolog (Drosophila) (218603_at), score: 0.71 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: 0.74 HIF1Ahypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (200989_at), score: 0.55 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: 0.61 HIST2H2BEhistone cluster 2, H2be (202708_s_at), score: 0.6 HIST3H2Ahistone cluster 3, H2a (221582_at), score: 0.54 HIVEP2human immunodeficiency virus type I enhancer binding protein 2 (212642_s_at), score: 0.56 HMGN4high mobility group nucleosomal binding domain 4 (209786_at), score: 0.54 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: -0.65 IFNGR1interferon gamma receptor 1 (211676_s_at), score: 0.54 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.85 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: 0.54 IL1Binterleukin 1, beta (39402_at), score: 0.59 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.62 IL1RNinterleukin 1 receptor antagonist (212657_s_at), score: 0.69 IL24interleukin 24 (206569_at), score: 0.72 ILF2interleukin enhancer binding factor 2, 45kDa (200052_s_at), score: -0.72 ING1inhibitor of growth family, member 1 (208415_x_at), score: 0.6 INSRinsulin receptor (213792_s_at), score: 0.76 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: 0.54 IRAK3interleukin-1 receptor-associated kinase 3 (213817_at), score: 0.63 ISCUiron-sulfur cluster scaffold homolog (E. coli) (209075_s_at), score: 0.72 JAK1Janus kinase 1 (a protein tyrosine kinase) (201648_at), score: 0.59 JMJD1Cjumonji domain containing 1C (221763_at), score: 0.74 KARSlysyl-tRNA synthetase (200079_s_at), score: -0.62 KCTD14potassium channel tetramerisation domain containing 14 (58916_at), score: -0.64 KIAA0247KIAA0247 (202181_at), score: 0.79 KIAA0831KIAA0831 (204568_at), score: 0.6 KIAA1324KIAA1324 (221874_at), score: -0.71 KIF18Bkinesin family member 18B (222039_at), score: -0.64 KLF10Kruppel-like factor 10 (202393_s_at), score: 0.54 KLF11Kruppel-like factor 11 (218486_at), score: 0.59 KLHDC2kelch domain containing 2 (217906_at), score: 0.64 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: -0.76 LACTB2lactamase, beta 2 (218701_at), score: 0.56 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: -0.67 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: -0.59 LEMD3LEM domain containing 3 (218604_at), score: 0.56 LEPREL1leprecan-like 1 (218717_s_at), score: -0.63 LGALS3lectin, galactoside-binding, soluble, 3 (208949_s_at), score: 0.54 LMAN2lectin, mannose-binding 2 (200805_at), score: -0.65 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: -0.6 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: 0.54 LOC220594TL132 protein (213510_x_at), score: 0.55 LOC728344similar to hCG1978918 (216532_x_at), score: -0.61 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.67 LRRC59leucine rich repeat containing 59 (222231_s_at), score: -0.67 LUMlumican (201744_s_at), score: 0.77 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.64 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: 0.7 MAN2C1mannosidase, alpha, class 2C, member 1 (203668_at), score: 0.53 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (218311_at), score: 0.62 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: -0.85 MARCH8membrane-associated ring finger (C3HC4) 8 (221824_s_at), score: 0.62 MBIPMAP3K12 binding inhibitory protein 1 (218411_s_at), score: 0.56 MED23mediator complex subunit 23 (218846_at), score: 0.56 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.63 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: 0.57 MICAL3microtubule associated monoxygenase, calponin and LIM domain containing 3 (212715_s_at), score: -0.59 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: 0.55 MOAP1modulator of apoptosis 1 (212508_at), score: 0.74 MORC3MORC family CW-type zinc finger 3 (213000_at), score: 0.67 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: -0.83 MPRIPmyosin phosphatase Rho interacting protein (212197_x_at), score: -0.6 MRPL24mitochondrial ribosomal protein L24 (218270_at), score: -0.65 MRPL40mitochondrial ribosomal protein L40 (203152_at), score: -0.63 MTHFD2Lmethylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2-like (220346_at), score: 0.56 MTHFS5,10-methenyltetrahydrofolate synthetase (5-formyltetrahydrofolate cyclo-ligase) (203433_at), score: -0.61 MXI1MAX interactor 1 (202364_at), score: 0.63 MYO1Bmyosin IB (212364_at), score: 0.79 NAMPTnicotinamide phosphoribosyltransferase (217738_at), score: 0.65 NAT15N-acetyltransferase 15 (GCN5-related, putative) (45526_g_at), score: 0.64 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: 0.55 NBPF12neuroblastoma breakpoint family, member 12 (213612_x_at), score: 0.59 NEIL3nei endonuclease VIII-like 3 (E. coli) (219502_at), score: -0.6 NFE2L3nuclear factor (erythroid-derived 2)-like 3 (204702_s_at), score: -0.85 NMT2N-myristoyltransferase 2 (205005_s_at), score: -0.68 NPAS2neuronal PAS domain protein 2 (39549_at), score: 0.63 NPM1nucleophosmin (nucleolar phosphoprotein B23, numatrin) (221923_s_at), score: -0.59 NPTNneuroplastin (202228_s_at), score: 0.62 NR4A2nuclear receptor subfamily 4, group A, member 2 (216248_s_at), score: 0.55 NR4A3nuclear receptor subfamily 4, group A, member 3 (209959_at), score: 0.6 NRD1nardilysin (N-arginine dibasic convertase) (208709_s_at), score: -0.63 NRP1neuropilin 1 (212298_at), score: 0.59 NUDCnuclear distribution gene C homolog (A. nidulans) (201173_x_at), score: -0.63 NUDCD3NudC domain containing 3 (201270_x_at), score: -0.62 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (212183_at), score: 0.62 NUMBnumb homolog (Drosophila) (207545_s_at), score: 0.61 OATornithine aminotransferase (gyrate atrophy) (201599_at), score: 0.56 OGTO-linked N-acetylglucosamine (GlcNAc) transferase (UDP-N-acetylglucosamine:polypeptide-N-acetylglucosaminyl transferase) (209240_at), score: 0.56 OLA1Obg-like ATPase 1 (219293_s_at), score: -0.65 OSBPL8oxysterol binding protein-like 8 (212582_at), score: 0.66 OSGIN2oxidative stress induced growth inhibitor family member 2 (204024_at), score: 0.52 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.66 PCBP2poly(rC) binding protein 2 (204031_s_at), score: 0.55 PCF11PCF11, cleavage and polyadenylation factor subunit, homolog (S. cerevisiae) (203378_at), score: 0.58 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: -0.93 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: 0.66 PCNXpecanex homolog (Drosophila) (213173_at), score: 0.53 PCTK2PCTAIRE protein kinase 2 (221918_at), score: 0.79 PCYOX1Lprenylcysteine oxidase 1 like (218953_s_at), score: -0.67 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.78 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: 0.63 PDZRN3PDZ domain containing ring finger 3 (212915_at), score: 0.54 PEG10paternally expressed 10 (212094_at), score: -0.61 PELI1pellino homolog 1 (Drosophila) (218319_at), score: 0.68 PELI2pellino homolog 2 (Drosophila) (219132_at), score: 0.67 PFN1profilin 1 (200634_at), score: -0.61 PHBprohibitin (200658_s_at), score: -0.59 PHF2PHD finger protein 2 (212726_at), score: 0.59 PHF21APHD finger protein 21A (203278_s_at), score: 0.52 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.62 PIP5K3phosphatidylinositol-3-phosphate/phosphatidylinositol 5-kinase, type III (213111_at), score: 0.65 PISDphosphatidylserine decarboxylase (202392_s_at), score: -0.63 PJA2praja ring finger 2 (201133_s_at), score: 0.72 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: 0.72 PKM2pyruvate kinase, muscle (201251_at), score: -0.63 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: 0.58 PLCG1phospholipase C, gamma 1 (202789_at), score: -0.59 PLCL2phospholipase C-like 2 (213309_at), score: 0.66 PLP2proteolipid protein 2 (colonic epithelium-enriched) (201136_at), score: -0.59 PNPOpyridoxamine 5'-phosphate oxidase (218511_s_at), score: -0.61 POLD2polymerase (DNA directed), delta 2, regulatory subunit 50kDa (201115_at), score: -0.67 POLR2Lpolymerase (RNA) II (DNA directed) polypeptide L, 7.6kDa (211730_s_at), score: -0.61 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (206653_at), score: -0.74 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: -0.61 PPP2R2Bprotein phosphatase 2 (formerly 2A), regulatory subunit B, beta isoform (213849_s_at), score: -0.86 PRDM2PR domain containing 2, with ZNF domain (203057_s_at), score: 0.61 PRDX1peroxiredoxin 1 (208680_at), score: -0.63 PRKAR2Bprotein kinase, cAMP-dependent, regulatory, type II, beta (203680_at), score: 0.53 PRPF39PRP39 pre-mRNA processing factor 39 homolog (S. cerevisiae) (220553_s_at), score: 0.63 PRR16proline rich 16 (220014_at), score: 0.6 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (205618_at), score: 0.64 PRRX1paired related homeobox 1 (205991_s_at), score: 0.68 PSMA7proteasome (prosome, macropain) subunit, alpha type, 7 (216088_s_at), score: -0.59 PSMB2proteasome (prosome, macropain) subunit, beta type, 2 (200039_s_at), score: -0.62 PSMB6proteasome (prosome, macropain) subunit, beta type, 6 (208827_at), score: -0.69 PSMD4proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 (211609_x_at), score: -0.67 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: 0.64 PTGESprostaglandin E synthase (210367_s_at), score: 0.65 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: 0.57 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: 0.53 PUM2pumilio homolog 2 (Drosophila) (216221_s_at), score: 0.67 PVRpoliovirus receptor (212662_at), score: -0.7 QPCTglutaminyl-peptide cyclotransferase (205174_s_at), score: 0.55 RAB21RAB21, member RAS oncogene family (203885_at), score: 0.53 RAB8ARAB8A, member RAS oncogene family (208819_at), score: -0.59 RABL4RAB, member of RAS oncogene family-like 4 (205037_at), score: -0.64 RAD51CRAD51 homolog C (S. cerevisiae) (209849_s_at), score: -0.64 RARBretinoic acid receptor, beta (205080_at), score: -0.71 RCN2reticulocalbin 2, EF-hand calcium binding domain (201485_s_at), score: 0.69 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.52 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (208070_s_at), score: 0.54 RGS5regulator of G-protein signaling 5 (218353_at), score: 0.65 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: -0.71 RND3Rho family GTPase 3 (212724_at), score: 0.81 RNF111ring finger protein 111 (218761_at), score: 0.6 RNF138ring finger protein 138 (218738_s_at), score: 0.52 RNF139ring finger protein 139 (209510_at), score: 0.64 RNF38ring finger protein 38 (218528_s_at), score: 0.56 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: 0.58 RPH3ALrabphilin 3A-like (without C2 domains) (221614_s_at), score: -0.59 RPL34ribosomal protein L34 (200026_at), score: 0.54 RPS5ribosomal protein S5 (200024_at), score: -0.59 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: -0.76 RREB1ras responsive element binding protein 1 (203704_s_at), score: 0.52 RSL24D1ribosomal L24 domain containing 1 (217915_s_at), score: 0.73 S100A10S100 calcium binding protein A10 (200872_at), score: -0.61 SAE1SUMO1 activating enzyme subunit 1 (217946_s_at), score: -0.6 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.75 SCAMP3secretory carrier membrane protein 3 (201771_at), score: -0.59 SCAMP4secretory carrier membrane protein 4 (213244_at), score: -0.6 SEC24BSEC24 family, member B (S. cerevisiae) (202798_at), score: 0.64 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: 0.56 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.75 SERINC1serine incorporator 1 (208671_at), score: 0.78 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.61 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.53 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: 0.58 SFRS5splicing factor, arginine/serine-rich 5 (212266_s_at), score: 0.69 SH2B3SH2B adaptor protein 3 (203320_at), score: 0.69 SH3BGRSH3 domain binding glutamic acid-rich protein (204979_s_at), score: -0.67 SHOC2soc-2 suppressor of clear homolog (C. elegans) (202777_at), score: 0.6 SIRT1sirtuin (silent mating type information regulation 2 homolog) 1 (S. cerevisiae) (218878_s_at), score: 0.63 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: -0.69 SLC19A1solute carrier family 19 (folate transporter), member 1 (211576_s_at), score: -0.65 SLC25A5solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 (200657_at), score: -0.71 SLC2A1solute carrier family 2 (facilitated glucose transporter), member 1 (201250_s_at), score: -0.75 SLC2A14solute carrier family 2 (facilitated glucose transporter), member 14 (222088_s_at), score: 0.6 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: 0.65 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: 0.67 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: 0.53 SLC35D2solute carrier family 35, member D2 (213082_s_at), score: -0.6 SLC7A5solute carrier family 7 (cationic amino acid transporter, y+ system), member 5 (201195_s_at), score: -0.61 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (204588_s_at), score: 0.61 SLKSTE20-like kinase (yeast) (206875_s_at), score: 0.81 SMAP1small ArfGAP 1 (218137_s_at), score: 0.64 SMARCE1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 (211989_at), score: -0.59 SMSspermine synthase (202043_s_at), score: -0.61 SNRPD3small nuclear ribonucleoprotein D3 polypeptide 18kDa (202567_at), score: -0.64 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: 0.73 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.81 SORDsorbitol dehydrogenase (201563_at), score: -0.62 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: 0.65 SOX4SRY (sex determining region Y)-box 4 (201417_at), score: 0.52 SPANXCSPANX family, member C (220217_x_at), score: -0.75 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: 0.7 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: 0.7 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: 0.54 STACSH3 and cysteine rich domain (205743_at), score: -0.67 STIM1stromal interaction molecule 1 (202764_at), score: -0.63 STK38Lserine/threonine kinase 38 like (212572_at), score: 0.63 STRN3striatin, calmodulin binding protein 3 (204496_at), score: 0.61 STX12syntaxin 12 (212112_s_at), score: 0.56 STYXL1serine/threonine/tyrosine interacting-like 1 (218321_x_at), score: -0.61 SVILsupervillin (202565_s_at), score: 0.65 SYPL1synaptophysin-like 1 (201260_s_at), score: 0.59 TBX2T-box 2 (40560_at), score: 0.59 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: 0.69 TEStestis derived transcript (3 LIM domains) (202720_at), score: 0.65 TFB2Mtranscription factor B2, mitochondrial (218605_at), score: 0.51 TGFB2transforming growth factor, beta 2 (220407_s_at), score: -0.6 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: -0.65 TGIF2TGFB-induced factor homeobox 2 (216262_s_at), score: -0.65 THBS2thrombospondin 2 (203083_at), score: 0.61 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (218408_at), score: -0.59 TIMM8Btranslocase of inner mitochondrial membrane 8 homolog B (yeast) (218357_s_at), score: -0.63 TIMP1TIMP metallopeptidase inhibitor 1 (201666_at), score: 0.56 TMED9transmembrane emp24 protein transport domain containing 9 (208757_at), score: -0.69 TMEM22transmembrane protein 22 (219569_s_at), score: 0.57 TMEM30Atransmembrane protein 30A (217743_s_at), score: 0.66 TMEM50Atransmembrane protein 50A (217766_s_at), score: 0.58 TMF1TATA element modulatory factor 1 (213024_at), score: 0.56 TNFAIP3tumor necrosis factor, alpha-induced protein 3 (202644_s_at), score: 0.56 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.55 TNS1tensin 1 (221748_s_at), score: 0.56 TOB1transducer of ERBB2, 1 (202704_at), score: 0.55 TOPORStopoisomerase I binding, arginine/serine-rich (204071_s_at), score: 0.55 TOR3Atorsin family 3, member A (218459_at), score: -0.65 TPBGtrophoblast glycoprotein (203476_at), score: 0.73 TPD52tumor protein D52 (201690_s_at), score: -0.69 TPM1tropomyosin 1 (alpha) (206116_s_at), score: -0.69 TRAPPC2trafficking protein particle complex 2 (219351_at), score: 0.54 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: 0.64 TRPV1transient receptor potential cation channel, subfamily V, member 1 (219632_s_at), score: 0.62 TSPYL4TSPY-like 4 (212928_at), score: 0.52 TSPYL5TSPY-like 5 (213122_at), score: 0.55 TTLL12tubulin tyrosine ligase-like family, member 12 (216251_s_at), score: -0.67 TUBA1Ctubulin, alpha 1c (211750_x_at), score: -0.68 TUBBtubulin, beta (209026_x_at), score: -0.73 TUBB2Ctubulin, beta 2C (213726_x_at), score: -0.62 TUBB3tubulin, beta 3 (213476_x_at), score: -0.59 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (211763_s_at), score: 0.57 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: 0.53 UBE2D3ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) (200669_s_at), score: 0.6 UBE2E1ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) (212519_at), score: 0.56 USP6NLUSP6 N-terminal like (204761_at), score: 0.59 VIMvimentin (201426_s_at), score: -0.6 WASF3WAS protein family, member 3 (204042_at), score: 0.8 WDR82WD repeat domain 82 (201934_at), score: 0.53 WWC1WW and C2 domain containing 1 (213085_s_at), score: -0.75 YAF2YY1 associated factor 2 (206238_s_at), score: 0.55 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.69 YOD1YOD1 OTU deubiquinating enzyme 1 homolog (S. cerevisiae) (215150_at), score: 0.53 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: 0.8 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: 0.54 ZBTB39zinc finger and BTB domain containing 39 (205256_at), score: 0.61 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.66 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: 0.72 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: 0.69 ZFAND5zinc finger, AN1-type domain 5 (217741_s_at), score: 0.7 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: 0.66 ZNF770zinc finger protein 770 (220608_s_at), score: 0.58
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956138.cel | 4 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956634.cel | 19 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |