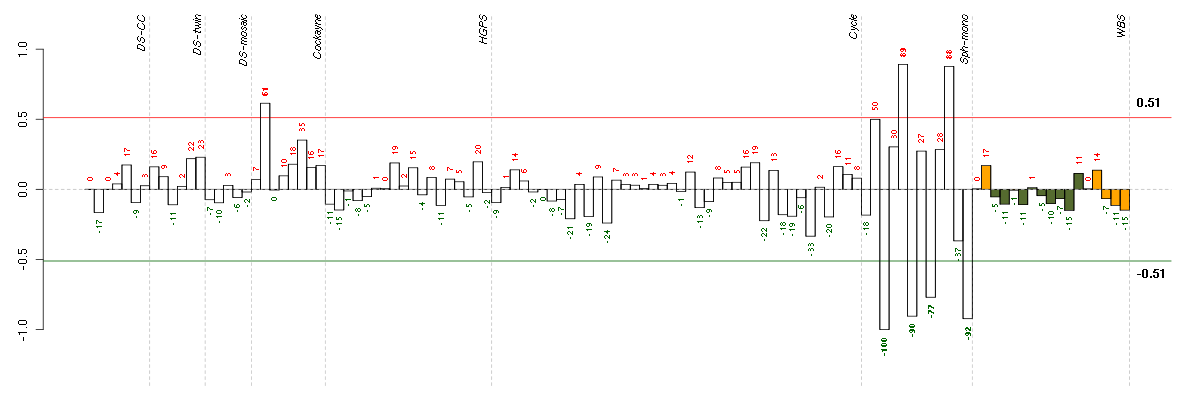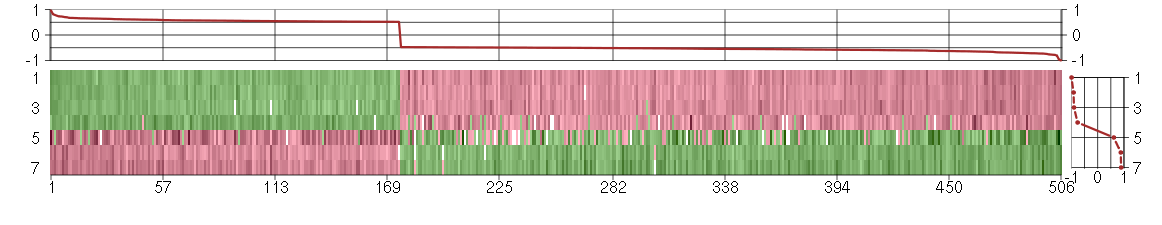

Under-expression is coded with green,
over-expression with red color.

modification-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent modification of the target protein.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
proteolysis
The hydrolysis of a peptide bond or bonds within a protein.
ubiquitin-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin moiety, or multiple ubiquitin moieties, to the protein.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
proteasomal protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds that is mediated by the proteasome.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer catabolic process
The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
proteasomal ubiquitin-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins.
biopolymer catabolic process
The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
modification-dependent macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, initiated by covalent modification of the target molecule.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein during the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
all
This term is the most general term possible
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
biopolymer catabolic process
The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature e.g. polysaccharides and proteins.
cellular biopolymer metabolic process
The chemical reactions and pathways involving biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
cellular biopolymer catabolic process
The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular biopolymer catabolic process
The chemical reactions and pathways resulting in the breakdown of biopolymers, long, repeating chains of monomers found in nature, such as polysaccharides and proteins, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein during the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
modification-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent modification of the target protein.
proteasomal ubiquitin-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome.


ABAT4-aminobutyrate aminotransferase (209459_s_at), score: -0.5 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: -0.5 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: -0.58 ABCC10ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (213485_s_at), score: -0.48 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.56 ADAM9ADAM metallopeptidase domain 9 (meltrin gamma) (202381_at), score: -0.54 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: -0.49 ADNPactivity-dependent neuroprotector homeobox (201773_at), score: -0.52 ADO2-aminoethanethiol (cysteamine) dioxygenase (212500_at), score: -0.52 AEBP1AE binding protein 1 (201792_at), score: -0.69 AGPSalkylglycerone phosphate synthase (205401_at), score: 0.63 AHI1Abelson helper integration site 1 (221569_at), score: -0.58 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: -0.61 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: -0.5 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.7 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: -0.61 ALDH3A2aldehyde dehydrogenase 3 family, member A2 (202053_s_at), score: 0.54 ANGangiogenin, ribonuclease, RNase A family, 5 (213397_x_at), score: -0.56 ANXA8L2annexin A8-like 2 (203074_at), score: 0.54 AP1S1adaptor-related protein complex 1, sigma 1 subunit (205195_at), score: 0.55 APH1Banterior pharynx defective 1 homolog B (C. elegans) (221036_s_at), score: -0.48 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (202629_at), score: -0.49 ARF4ADP-ribosylation factor 4 (201096_s_at), score: -0.62 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.54 ARFIP1ADP-ribosylation factor interacting protein 1 (218230_at), score: -0.55 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: -0.7 ARHGAP6Rho GTPase activating protein 6 (206167_s_at), score: -0.49 ARID4AAT rich interactive domain 4A (RBP1-like) (205062_x_at), score: -0.56 ARID5BAT rich interactive domain 5B (MRF1-like) (212614_at), score: -0.55 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.65 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.7 ARMCX3armadillo repeat containing, X-linked 3 (217858_s_at), score: -0.51 ARSJarylsulfatase family, member J (219973_at), score: -0.48 ASAP2ArfGAP with SH3 domain, ankyrin repeat and PH domain 2 (206414_s_at), score: -0.59 ASPNasporin (219087_at), score: -0.49 ATF1activating transcription factor 1 (222103_at), score: -0.6 ATF7IP2activating transcription factor 7 interacting protein 2 (219870_at), score: -0.57 ATMINATM interactor (201855_s_at), score: -0.49 ATP1A1ATPase, Na+/K+ transporting, alpha 1 polypeptide (220948_s_at), score: 0.63 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (212135_s_at), score: -0.51 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.57 AUP1ancient ubiquitous protein 1 (220525_s_at), score: 0.6 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: -0.63 BEX4brain expressed, X-linked 4 (215440_s_at), score: -0.52 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: -0.56 BIRC2baculoviral IAP repeat-containing 2 (202076_at), score: 0.53 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: 0.63 BMP7bone morphogenetic protein 7 (209590_at), score: 0.52 C10orf116chromosome 10 open reading frame 116 (203571_s_at), score: -0.54 C10orf18chromosome 10 open reading frame 18 (218331_s_at), score: -0.53 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.51 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.64 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: -0.59 C16orf57chromosome 16 open reading frame 57 (218060_s_at), score: 0.59 C16orf88chromosome 16 open reading frame 88 (213237_at), score: 0.52 C1QBPcomplement component 1, q subcomponent binding protein (208910_s_at), score: 0.53 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.53 C21orf59chromosome 21 open reading frame 59 (218123_at), score: 0.55 C2orf37chromosome 2 open reading frame 37 (220172_at), score: -0.49 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.48 C5orf4chromosome 5 open reading frame 4 (48031_r_at), score: -0.65 C6orf120chromosome 6 open reading frame 120 (221786_at), score: -0.53 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.55 C6orf47chromosome 6 open reading frame 47 (204968_at), score: 0.55 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.64 C9orf82chromosome 9 open reading frame 82 (219276_x_at), score: -0.53 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: -0.55 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.64 CCDC59coiled-coil domain containing 59 (218936_s_at), score: 0.52 CCL2chemokine (C-C motif) ligand 2 (216598_s_at), score: -0.5 CCND3cyclin D3 (201700_at), score: 0.53 CCNG1cyclin G1 (208796_s_at), score: -0.57 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.54 CD24CD24 molecule (209771_x_at), score: 0.64 CD248CD248 molecule, endosialin (219025_at), score: -0.62 CD302CD302 molecule (203799_at), score: -0.5 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.55 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: 0.53 CDK5RAP1CDK5 regulatory subunit associated protein 1 (218315_s_at), score: 0.58 CDKN1Acyclin-dependent kinase inhibitor 1A (p21, Cip1) (202284_s_at), score: -0.62 CELSR1cadherin, EGF LAG seven-pass G-type receptor 1 (flamingo homolog, Drosophila) (41660_at), score: 0.56 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.82 CENPJcentromere protein J (220885_s_at), score: 0.52 CEP170centrosomal protein 170kDa (212746_s_at), score: -0.52 CFHcomplement factor H (213800_at), score: -0.53 CHCHD2coiled-coil-helix-coiled-coil-helix domain containing 2 (217720_at), score: 0.54 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.66 CHMP7CHMP family, member 7 (212313_at), score: 0.52 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.55 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.52 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: -0.48 CNIHcornichon homolog (Drosophila) (201653_at), score: -0.55 CNNM1cyclin M1 (220166_at), score: -0.48 COBLL1COBL-like 1 (203642_s_at), score: -0.49 COL15A1collagen, type XV, alpha 1 (203477_at), score: -0.5 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.69 CPEB3cytoplasmic polyadenylation element binding protein 3 (205773_at), score: -0.54 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (206688_s_at), score: 0.57 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.6 CTNNBL1catenin, beta like 1 (221021_s_at), score: 0.61 CTSKcathepsin K (202450_s_at), score: -0.48 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.6 CYB5R2cytochrome b5 reductase 2 (220230_s_at), score: -0.59 DAPK1death-associated protein kinase 1 (203139_at), score: -0.58 DCNdecorin (211896_s_at), score: -0.61 DCUN1D1DCN1, defective in cullin neddylation 1, domain containing 1 (S. cerevisiae) (218583_s_at), score: -0.49 DERL1Der1-like domain family, member 1 (219402_s_at), score: 0.61 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.75 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.56 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.64 DPY19L4dpy-19-like 4 (C. elegans) (213391_at), score: -0.58 DPYSL2dihydropyrimidinase-like 2 (200762_at), score: -0.52 DRAP1DR1-associated protein 1 (negative cofactor 2 alpha) (203258_at), score: 0.6 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: 0.66 DSEdermatan sulfate epimerase (218854_at), score: -0.64 DSPdesmoplakin (200606_at), score: -0.49 DZIP1DAZ interacting protein 1 (204557_s_at), score: -0.52 E2F1E2F transcription factor 1 (204947_at), score: 0.6 EDNRAendothelin receptor type A (204464_s_at), score: -0.48 EEF1Geukaryotic translation elongation factor 1 gamma (200689_x_at), score: 0.53 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.53 EIF2C2eukaryotic translation initiation factor 2C, 2 (213310_at), score: -0.6 EIF3Deukaryotic translation initiation factor 3, subunit D (200005_at), score: 0.64 EIF4A1eukaryotic translation initiation factor 4A, isoform 1 (211787_s_at), score: 0.66 EIF4A3eukaryotic translation initiation factor 4A, isoform 3 (201303_at), score: 0.56 EIF6eukaryotic translation initiation factor 6 (210213_s_at), score: 0.62 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: -0.58 EMP1epithelial membrane protein 1 (201325_s_at), score: -0.63 ENOSF1enolase superfamily member 1 (204142_at), score: -0.54 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: -0.6 EPAS1endothelial PAS domain protein 1 (200878_at), score: -0.52 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.61 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.63 EREGepiregulin (205767_at), score: -0.49 ERI3exoribonuclease 3 (208973_at), score: 0.54 ESF1ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) (218859_s_at), score: 0.52 EWSR1Ewing sarcoma breakpoint region 1 (209214_s_at), score: 0.55 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.57 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.78 FAM46Cfamily with sequence similarity 46, member C (220306_at), score: -0.48 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: -0.66 FANCAFanconi anemia, complementation group A (203805_s_at), score: 0.52 FBN2fibrillin 2 (203184_at), score: -0.54 FCHSD2FCH and double SH3 domains 2 (203620_s_at), score: -0.57 FKBP11FK506 binding protein 11, 19 kDa (219118_at), score: -0.53 FLJ10357hypothetical protein FLJ10357 (58780_s_at), score: -0.53 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: -0.67 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.65 FOSL1FOS-like antigen 1 (204420_at), score: 0.57 FOXG1forkhead box G1 (206018_at), score: 0.62 FOXL2forkhead box L2 (220102_at), score: 0.62 FOXO1forkhead box O1 (202724_s_at), score: -0.5 FYCO1FYVE and coiled-coil domain containing 1 (218204_s_at), score: -0.49 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: -0.5 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.69 GALCgalactosylceramidase (204417_at), score: -0.72 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: -0.5 GAS1growth arrest-specific 1 (204457_s_at), score: -0.58 GCC2GRIP and coiled-coil domain containing 2 (202832_at), score: -0.54 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: -0.48 GIPC2GIPC PDZ domain containing family, member 2 (219970_at), score: -0.55 GLAgalactosidase, alpha (214430_at), score: 0.55 GLRX3glutaredoxin 3 (209080_x_at), score: 0.55 GLT25D2glycosyltransferase 25 domain containing 2 (209883_at), score: -0.49 GNG12guanine nucleotide binding protein (G protein), gamma 12 (212294_at), score: -0.6 GOLPH3golgi phosphoprotein 3 (coat-protein) (217803_at), score: -0.58 GPR126G protein-coupled receptor 126 (213094_at), score: -0.58 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.49 GPR56G protein-coupled receptor 56 (212070_at), score: 0.56 GRAMD3GRAM domain containing 3 (218706_s_at), score: -0.56 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 GRIK2glutamate receptor, ionotropic, kainate 2 (213845_at), score: -0.52 GRINL1Aglutamate receptor, ionotropic, N-methyl D-aspartate-like 1A (212244_at), score: -0.5 HDAC3histone deacetylase 3 (216326_s_at), score: 0.52 HIF1Ahypoxia inducible factor 1, alpha subunit (basic helix-loop-helix transcription factor) (200989_at), score: -0.56 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.5 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: -0.49 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: 0.54 HMGB3L1high-mobility group box 3-like 1 (216548_x_at), score: 0.55 HOXA10homeobox A10 (213150_at), score: -0.6 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: -0.72 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.52 HSP90AB1heat shock protein 90kDa alpha (cytosolic), class B member 1 (214359_s_at), score: 0.53 HYPKHuntingtin interacting protein K (218680_x_at), score: 0.73 ICT1immature colon carcinoma transcript 1 (204868_at), score: 0.52 IDH3Bisocitrate dehydrogenase 3 (NAD+) beta (210014_x_at), score: 0.52 IFIT5interferon-induced protein with tetratricopeptide repeats 5 (203596_s_at), score: -0.5 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.79 IKIK cytokine, down-regulator of HLA II (200066_at), score: 0.57 IL10RBinterleukin 10 receptor, beta (209575_at), score: 0.54 IL1R1interleukin 1 receptor, type I (202948_at), score: -0.57 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.66 ILF2interleukin enhancer binding factor 2, 45kDa (200052_s_at), score: 0.57 IMPACTImpact homolog (mouse) (218637_at), score: -0.49 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.49 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: -0.69 IRAK3interleukin-1 receptor-associated kinase 3 (213817_at), score: -0.53 ISL1ISL LIM homeobox 1 (206104_at), score: -0.65 JAM3junctional adhesion molecule 3 (212813_at), score: -0.48 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.48 KARSlysyl-tRNA synthetase (200079_s_at), score: 0.58 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: -0.49 KCNN2potassium intermediate/small conductance calcium-activated channel, subfamily N, member 2 (220116_at), score: -0.5 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.52 KIAA1128KIAA1128 (209379_s_at), score: -0.58 KIAA1199KIAA1199 (212942_s_at), score: -0.48 KIF3Bkinesin family member 3B (203943_at), score: 0.54 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.96 LAMB3laminin, beta 3 (209270_at), score: 0.57 LAPTM5lysosomal multispanning membrane protein 5 (201721_s_at), score: 0.52 LAS1LLAS1-like (S. cerevisiae) (208117_s_at), score: 0.63 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.67 LGMNlegumain (201212_at), score: -0.54 LIPAlipase A, lysosomal acid, cholesterol esterase (201847_at), score: -0.61 LMBRD1LMBR1 domain containing 1 (218191_s_at), score: -0.52 LMO7LIM domain 7 (202674_s_at), score: -0.55 LOC100129250similar to hCG1811779 (78383_at), score: -0.49 LOC220594TL132 protein (213510_x_at), score: -0.51 LOC728344similar to hCG1978918 (216532_x_at), score: 0.62 LOC728855hypothetical LOC728855 (222001_x_at), score: 0.64 LPPR4plasticity related gene 1 (213496_at), score: -0.7 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: -0.59 LRRC2leucine rich repeat containing 2 (219949_at), score: -0.63 LSG1large subunit GTPase 1 homolog (S. cerevisiae) (221536_s_at), score: 0.57 LTBP2latent transforming growth factor beta binding protein 2 (204682_at), score: -0.62 LUMlumican (201744_s_at), score: -0.97 LXNlatexin (218729_at), score: -0.64 MANSC1MANSC domain containing 1 (220945_x_at), score: -0.52 MAPRE3microtubule-associated protein, RP/EB family, member 3 (203842_s_at), score: 0.57 MARCH2membrane-associated ring finger (C3HC4) 2 (210075_at), score: -0.48 MASP1mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) (213749_at), score: -0.53 MBNL1muscleblind-like (Drosophila) (201151_s_at), score: -0.49 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204058_at), score: -0.51 MED23mediator complex subunit 23 (218846_at), score: -0.64 MED9mediator complex subunit 9 (218372_at), score: 0.52 MEIS1Meis homeobox 1 (204069_at), score: -0.51 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: -0.73 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: -0.51 MLF1myeloid leukemia factor 1 (204784_s_at), score: 0.55 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: -0.68 MMEmembrane metallo-endopeptidase (203434_s_at), score: -0.49 MORC3MORC family CW-type zinc finger 3 (213000_at), score: -0.55 MOXD1monooxygenase, DBH-like 1 (209708_at), score: -0.62 MPPED2metallophosphoesterase domain containing 2 (205413_at), score: 0.65 MRPL22mitochondrial ribosomal protein L22 (218339_at), score: 0.52 MRPL28mitochondrial ribosomal protein L28 (204599_s_at), score: 0.57 MRPL9mitochondrial ribosomal protein L9 (211594_s_at), score: 0.57 MRPS18Cmitochondrial ribosomal protein S18C (220103_s_at), score: -0.51 MRPS7mitochondrial ribosomal protein S7 (217932_at), score: 0.53 MTDHmetadherin (212251_at), score: 0.54 MUDENGMU-2/AP1M2 domain containing, death-inducing (218139_s_at), score: -0.49 MXRA5matrix-remodelling associated 5 (209596_at), score: -0.56 MXRA8matrix-remodelling associated 8 (213422_s_at), score: -0.48 MYO1Bmyosin IB (212364_at), score: -0.78 MYO6myosin VI (203216_s_at), score: -0.55 MYST4MYST histone acetyltransferase (monocytic leukemia) 4 (212462_at), score: -0.51 NASPnuclear autoantigenic sperm protein (histone-binding) (201970_s_at), score: 0.65 NBEAneurobeachin (221207_s_at), score: -0.59 NDNnecdin homolog (mouse) (209550_at), score: -0.53 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (201740_at), score: 0.52 NEK7NIMA (never in mitosis gene a)-related kinase 7 (212530_at), score: -0.64 NESnestin (218678_at), score: -0.6 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: -0.5 NIP7nuclear import 7 homolog (S. cerevisiae) (219031_s_at), score: 0.53 NLGN1neuroligin 1 (205893_at), score: -0.57 NOL12nucleolar protein 12 (219324_at), score: 0.52 NOP16NOP16 nucleolar protein homolog (yeast) (203023_at), score: 0.54 NPAS2neuronal PAS domain protein 2 (39549_at), score: -0.59 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (217796_s_at), score: 0.6 NPTNneuroplastin (202228_s_at), score: -0.54 NPTX1neuronal pentraxin I (204684_at), score: 0.6 NRP1neuropilin 1 (212298_at), score: -0.56 NSFL1CNSFL1 (p97) cofactor (p47) (217831_s_at), score: 0.6 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: -0.51 NUDCnuclear distribution gene C homolog (A. nidulans) (201173_x_at), score: 0.53 NUDT2nudix (nucleoside diphosphate linked moiety X)-type motif 2 (218609_s_at), score: 0.53 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (212183_at), score: -0.52 NUMBnumb homolog (Drosophila) (207545_s_at), score: -0.49 OATornithine aminotransferase (gyrate atrophy) (201599_at), score: -0.5 OLFML3olfactomedin-like 3 (218162_at), score: -0.49 OPTNoptineurin (202073_at), score: -0.58 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: -0.58 OSBPL2oxysterol binding protein-like 2 (209222_s_at), score: 0.54 OXTRoxytocin receptor (206825_at), score: -0.53 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: -0.58 PBX1pre-B-cell leukemia homeobox 1 (212148_at), score: -0.56 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: -0.63 PCGF1polycomb group ring finger 1 (210023_s_at), score: 0.52 PCLOpiccolo (presynaptic cytomatrix protein) (213558_at), score: 0.72 PCMTD2protein-L-isoaspartate (D-aspartate) O-methyltransferase domain containing 2 (212406_s_at), score: -0.49 PCNXpecanex homolog (Drosophila) (213173_at), score: -0.55 PCTK2PCTAIRE protein kinase 2 (221918_at), score: -0.74 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.77 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: -0.5 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.57 PFDN2prefoldin subunit 2 (218336_at), score: 0.73 PFDN6prefoldin subunit 6 (222029_x_at), score: 0.53 PGAP1post-GPI attachment to proteins 1 (213469_at), score: -0.63 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.56 PJA2praja ring finger 2 (201133_s_at), score: -0.73 PKD2polycystic kidney disease 2 (autosomal dominant) (203688_at), score: -0.57 PKIGprotein kinase (cAMP-dependent, catalytic) inhibitor gamma (202732_at), score: -0.57 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.61 PLAGL1pleiomorphic adenoma gene-like 1 (209318_x_at), score: -0.58 PLATplasminogen activator, tissue (201860_s_at), score: -0.48 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: -0.69 PLCG1phospholipase C, gamma 1 (202789_at), score: 0.62 PLK2polo-like kinase 2 (Drosophila) (201939_at), score: 0.56 PLSCR4phospholipid scramblase 4 (218901_at), score: -0.5 PMPCApeptidase (mitochondrial processing) alpha (212088_at), score: 0.54 POFUT1protein O-fucosyltransferase 1 (212349_at), score: 0.55 POLD2polymerase (DNA directed), delta 2, regulatory subunit 50kDa (201115_at), score: 0.56 POLR2Fpolymerase (RNA) II (DNA directed) polypeptide F (209511_at), score: 0.6 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.6 POSTNperiostin, osteoblast specific factor (210809_s_at), score: -0.55 PPIEpeptidylprolyl isomerase E (cyclophilin E) (210502_s_at), score: 0.56 PPIHpeptidylprolyl isomerase H (cyclophilin H) (204228_at), score: 0.65 PPP1R3Dprotein phosphatase 1, regulatory (inhibitor) subunit 3D (204554_at), score: -0.5 PQBP1polyglutamine binding protein 1 (214527_s_at), score: 0.58 PRCCpapillary renal cell carcinoma (translocation-associated) (208938_at), score: 0.52 PRR16proline rich 16 (220014_at), score: -0.66 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (205618_at), score: -0.54 PRRX1paired related homeobox 1 (205991_s_at), score: -0.52 PSMB1proteasome (prosome, macropain) subunit, beta type, 1 (200876_s_at), score: 0.58 PSMB3proteasome (prosome, macropain) subunit, beta type, 3 (201400_at), score: 0.54 PSMB6proteasome (prosome, macropain) subunit, beta type, 6 (208827_at), score: 0.65 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.58 PSMC3proteasome (prosome, macropain) 26S subunit, ATPase, 3 (201267_s_at), score: 0.55 PSMD11proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 (208777_s_at), score: 0.56 PSMD13proteasome (prosome, macropain) 26S subunit, non-ATPase, 13 (201232_s_at), score: 0.55 PSMD4proteasome (prosome, macropain) 26S subunit, non-ATPase, 4 (211609_x_at), score: 0.64 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: -0.55 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.76 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.51 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.48 PTPN1protein tyrosine phosphatase, non-receptor type 1 (202716_at), score: 0.56 PTPN13protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (204201_s_at), score: -0.49 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.71 PUM2pumilio homolog 2 (Drosophila) (216221_s_at), score: -0.52 PYROXD1pyridine nucleotide-disulphide oxidoreductase domain 1 (213878_at), score: -0.48 R3HDM2R3H domain containing 2 (203831_at), score: -0.58 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.61 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.48 RAD23BRAD23 homolog B (S. cerevisiae) (201222_s_at), score: -0.49 RALYRNA binding protein, autoantigenic (hnRNP-associated with lethal yellow homolog (mouse)) (201271_s_at), score: 0.57 RASL11BRAS-like, family 11, member B (219142_at), score: -0.48 RCN2reticulocalbin 2, EF-hand calcium binding domain (201485_s_at), score: -0.52 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.71 REV3LREV3-like, catalytic subunit of DNA polymerase zeta (yeast) (208070_s_at), score: -0.49 RGP1RGP1 retrograde golgi transport homolog (S. cerevisiae) (203169_at), score: 0.58 RGS4regulator of G-protein signaling 4 (204337_at), score: -0.68 RHOFras homolog gene family, member F (in filopodia) (219045_at), score: -0.48 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.65 RND3Rho family GTPase 3 (212724_at), score: -0.59 RNF111ring finger protein 111 (218761_at), score: -0.5 RNF121ring finger protein 121 (219021_at), score: 0.61 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (213044_at), score: -0.58 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.6 RPL22ribosomal protein L22 (221726_at), score: -0.52 RPP14ribonuclease P/MRP 14kDa subunit (204245_s_at), score: 0.52 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: -0.56 RPS20ribosomal protein S20 (216247_at), score: -0.49 RPS3ribosomal protein S3 (208692_at), score: 0.61 RPS4P17ribosomal protein S4X pseudogene 17 (216342_x_at), score: 0.56 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: 0.57 RPS5ribosomal protein S5 (200024_at), score: 0.52 RPS6KA1ribosomal protein S6 kinase, 90kDa, polypeptide 1 (203379_at), score: 0.59 RPS6KA2ribosomal protein S6 kinase, 90kDa, polypeptide 2 (212912_at), score: -0.5 RSL1D1ribosomal L1 domain containing 1 (212018_s_at), score: 0.58 RUNX1runt-related transcription factor 1 (209360_s_at), score: -0.59 S100A4S100 calcium binding protein A4 (203186_s_at), score: -0.51 SACSspastic ataxia of Charlevoix-Saguenay (sacsin) (213262_at), score: -0.49 SALL2sal-like 2 (Drosophila) (213283_s_at), score: -0.59 SAMD4Asterile alpha motif domain containing 4A (212845_at), score: -0.53 SAP30BPSAP30 binding protein (217965_s_at), score: 0.58 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: -0.54 SCRIBscribbled homolog (Drosophila) (212556_at), score: -0.52 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: -0.51 SDHBsuccinate dehydrogenase complex, subunit B, iron sulfur (Ip) (202675_at), score: 0.52 SEC24DSEC24 family, member D (S. cerevisiae) (202375_at), score: -0.53 SELTselenoprotein T (217811_at), score: -0.48 SEMA3Fsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F (35666_at), score: -0.61 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: -0.5 SERINC1serine incorporator 1 (208671_at), score: -0.51 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: -0.52 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.69 SESN1sestrin 1 (218346_s_at), score: -0.48 SF3A3splicing factor 3a, subunit 3, 60kDa (203818_s_at), score: 0.55 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: -0.55 SH3YL1SH3 domain containing, Ysc84-like 1 (S. cerevisiae) (204019_s_at), score: -0.5 SHOC2soc-2 suppressor of clear homolog (C. elegans) (202777_at), score: -0.52 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: -0.54 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: 0.54 SLC25A5solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 5 (200657_at), score: 0.57 SLC25A6solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 (212085_at), score: 0.52 SLC2A10solute carrier family 2 (facilitated glucose transporter), member 10 (221024_s_at), score: -0.55 SLC2A3P1solute carrier family 2 (facilitated glucose transporter), member 3 pseudogene 1 (221751_at), score: -0.5 SLC30A1solute carrier family 30 (zinc transporter), member 1 (212907_at), score: -0.65 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.52 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.6 SLC39A4solute carrier family 39 (zinc transporter), member 4 (219215_s_at), score: -0.51 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: -0.59 SMA4glucuronidase, beta pseudogene (215599_at), score: 0.57 SMA5glucuronidase, beta pseudogene (215043_s_at), score: 0.57 SMAD4SMAD family member 4 (202527_s_at), score: -0.57 SMAP1small ArfGAP 1 (218137_s_at), score: -0.57 SMARCE1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily e, member 1 (211989_at), score: 0.65 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: 0.67 SNRNP40small nuclear ribonucleoprotein 40kDa (U5) (215905_s_at), score: 0.55 SNRPA1small nuclear ribonucleoprotein polypeptide A' (216977_x_at), score: 0.54 SNRPBsmall nuclear ribonucleoprotein polypeptides B and B1 (213175_s_at), score: 0.53 SOCS5suppressor of cytokine signaling 5 (209647_s_at), score: -0.61 SOS2son of sevenless homolog 2 (Drosophila) (212870_at), score: -0.5 SPANXCSPANX family, member C (220217_x_at), score: 0.61 SPATA7spermatogenesis associated 7 (219583_s_at), score: -0.48 SPCS3signal peptidase complex subunit 3 homolog (S. cerevisiae) (218817_at), score: -0.5 SPHARS-phase response (cyclin-related) (206272_at), score: -0.63 SSPNsarcospan (Kras oncogene-associated gene) (204963_at), score: -0.55 SSRP1structure specific recognition protein 1 (200956_s_at), score: 0.67 ST3GAL5ST3 beta-galactoside alpha-2,3-sialyltransferase 5 (203217_s_at), score: -0.49 STAM2signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 (209649_at), score: -0.66 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: -0.49 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: -0.55 STIM1stromal interaction molecule 1 (202764_at), score: 0.53 STK38Lserine/threonine kinase 38 like (212572_at), score: -0.56 STOMstomatin (201060_x_at), score: -0.55 STOML2stomatin (EPB72)-like 2 (215416_s_at), score: 0.54 STRN3striatin, calmodulin binding protein 3 (204496_at), score: -0.54 STX12syntaxin 12 (212112_s_at), score: -0.52 STX7syntaxin 7 (212631_at), score: -0.55 SULF1sulfatase 1 (212353_at), score: -0.68 SULT1A1sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 (203615_x_at), score: 0.52 SYNGR1synaptogyrin 1 (210613_s_at), score: -0.71 SYT11synaptotagmin XI (209198_s_at), score: -0.51 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: -0.59 TBXA2Rthromboxane A2 receptor (336_at), score: -0.69 TCF12transcription factor 12 (208986_at), score: -0.57 TCF7L2transcription factor 7-like 2 (T-cell specific, HMG-box) (212761_at), score: -0.64 TEAD4TEA domain family member 4 (41037_at), score: 0.53 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.81 TFIP11tuftelin interacting protein 11 (202750_s_at), score: 0.6 TGFB2transforming growth factor, beta 2 (220407_s_at), score: 0.61 TGFBItransforming growth factor, beta-induced, 68kDa (201506_at), score: 0.56 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: -0.55 THBS2thrombospondin 2 (203083_at), score: -0.73 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: -0.5 TMEM111transmembrane protein 111 (217882_at), score: 0.61 TMEM135transmembrane protein 135 (222209_s_at), score: -0.49 TMEM168transmembrane protein 168 (218962_s_at), score: -0.58 TMEM186transmembrane protein 186 (204676_at), score: 0.54 TMEM2transmembrane protein 2 (218113_at), score: -0.55 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.72 TMEM50Atransmembrane protein 50A (217766_s_at), score: -0.54 TMEM80transmembrane protein 80 (65630_at), score: 0.54 TNS1tensin 1 (221748_s_at), score: -0.74 TOB1transducer of ERBB2, 1 (202704_at), score: -0.58 TOR3Atorsin family 3, member A (218459_at), score: 0.57 TPBGtrophoblast glycoprotein (203476_at), score: -0.48 TRAPPC2trafficking protein particle complex 2 (219351_at), score: -0.52 TRMUtRNA 5-methylaminomethyl-2-thiouridylate methyltransferase (213634_s_at), score: 0.52 TRNAG6transfer RNA glycine 6 (anticodon GCC) (217542_at), score: -0.51 TRNAU1APtRNA selenocysteine 1 associated protein 1 (218977_s_at), score: 0.53 TRPC3transient receptor potential cation channel, subfamily C, member 3 (210814_at), score: 0.7 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: -0.59 TSFMTs translation elongation factor, mitochondrial (212656_at), score: 0.52 TSPAN13tetraspanin 13 (217979_at), score: -0.58 TSPAN6tetraspanin 6 (209108_at), score: -0.56 TSPYL1TSPY-like 1 (221493_at), score: -0.54 TSPYL4TSPY-like 4 (212928_at), score: -0.51 TSPYL5TSPY-like 5 (213122_at), score: -0.72 TTC33tetratricopeptide repeat domain 33 (219421_at), score: -0.53 TTLL12tubulin tyrosine ligase-like family, member 12 (216251_s_at), score: 0.59 TYRP1tyrosinase-related protein 1 (205694_at), score: 0.54 U2AF1U2 small nuclear RNA auxiliary factor 1 (202858_at), score: 0.64 UBE2Bubiquitin-conjugating enzyme E2B (RAD6 homolog) (211763_s_at), score: -0.48 UBE2E1ubiquitin-conjugating enzyme E2E 1 (UBC4/5 homolog, yeast) (212519_at), score: -0.51 UBE2G2ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) (209042_s_at), score: 0.56 UBE2Iubiquitin-conjugating enzyme E2I (UBC9 homolog, yeast) (213535_s_at), score: 0.53 UBE2Subiquitin-conjugating enzyme E2S (202779_s_at), score: 0.57 UBR2ubiquitin protein ligase E3 component n-recognin 2 (212760_at), score: -0.48 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: -0.63 URODuroporphyrinogen decarboxylase (208971_at), score: 0.56 UTP18UTP18, small subunit (SSU) processome component, homolog (yeast) (203721_s_at), score: 0.65 UTYubiquitously transcribed tetratricopeptide repeat gene, Y-linked (211149_at), score: -0.64 VAV3vav 3 guanine nucleotide exchange factor (218807_at), score: 0.53 VGLL3vestigial like 3 (Drosophila) (220327_at), score: -0.63 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.55 WASF3WAS protein family, member 3 (204042_at), score: -0.49 WDR44WD repeat domain 44 (219297_at), score: -0.51 WDR74WD repeat domain 74 (221712_s_at), score: 0.52 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: -0.49 WWC1WW and C2 domain containing 1 (213085_s_at), score: 0.54 YBX1Y box binding protein 1 (208627_s_at), score: 0.53 YIPF4Yip1 domain family, member 4 (209551_at), score: -0.57 YPEL5yippee-like 5 (Drosophila) (217783_s_at), score: -0.7 YTHDF3YTH domain family, member 3 (221749_at), score: -0.48 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.52 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: -0.54 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: -0.55 ZDHHC17zinc finger, DHHC-type containing 17 (212982_at), score: -0.65 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.6 ZNF215zinc finger protein 215 (220214_at), score: -0.59 ZNF394zinc finger protein 394 (214714_at), score: 0.56 ZNF516zinc finger protein 516 (203604_at), score: -0.49 ZNF518Azinc finger protein 518A (204291_at), score: -0.6 ZNF770zinc finger protein 770 (220608_s_at), score: -0.51
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-3407-raw-cel-1437949579.cel | 2 | 4 | Cockayne | hgu133a | none | CSB |
| E-GEOD-4219-raw-cel-1311956614.cel | 18 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956321.cel | 9 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |