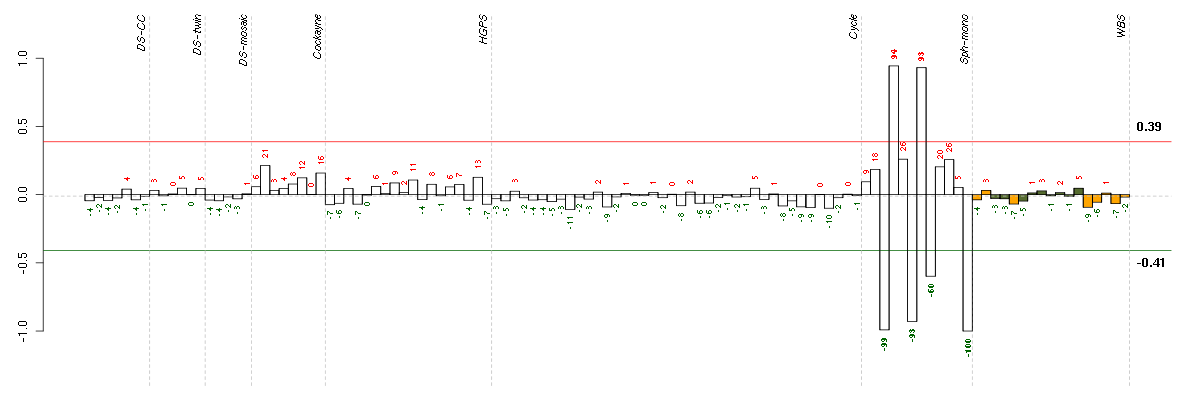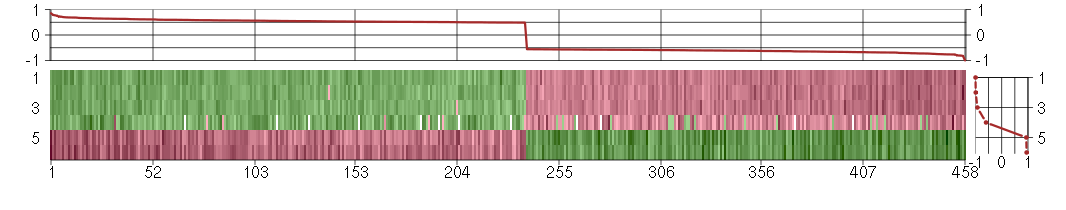

Under-expression is coded with green,
over-expression with red color.



ABCC10ATP-binding cassette, sub-family C (CFTR/MRP), member 10 (213485_s_at), score: -0.57 ABHD2abhydrolase domain containing 2 (63825_at), score: 0.55 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.64 ACO1aconitase 1, soluble (207071_s_at), score: -0.69 ACOX3acyl-Coenzyme A oxidase 3, pristanoyl (204241_at), score: -0.57 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.54 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: 0.58 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.59 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (214913_at), score: -0.71 ADIPOR1adiponectin receptor 1 (217748_at), score: 0.76 AEBP1AE binding protein 1 (201792_at), score: -0.6 AFTPHaftiphilin (217939_s_at), score: 0.51 AKAP12A kinase (PRKA) anchor protein 12 (210517_s_at), score: -0.58 ALDH1A1aldehyde dehydrogenase 1 family, member A1 (212224_at), score: -0.68 ALG5asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae) (218203_at), score: 0.52 ANAPC10anaphase promoting complex subunit 10 (207845_s_at), score: 0.49 ANKHD1ankyrin repeat and KH domain containing 1 (208772_at), score: 0.54 ARF5ADP-ribosylation factor 5 (201526_at), score: 0.51 ARFIP1ADP-ribosylation factor interacting protein 1 (218230_at), score: -0.58 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.72 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.57 ARMCX1armadillo repeat containing, X-linked 1 (218694_at), score: -0.69 ARMCX6armadillo repeat containing, X-linked 6 (214749_s_at), score: -0.61 ARSJarylsulfatase family, member J (219973_at), score: -0.61 ASCC3activating signal cointegrator 1 complex subunit 3 (212815_at), score: -0.65 ATG2BATG2 autophagy related 2 homolog B (S. cerevisiae) (219164_s_at), score: -0.67 ATG3ATG3 autophagy related 3 homolog (S. cerevisiae) (221492_s_at), score: 0.52 ATP5G2ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit 9) (208764_s_at), score: -0.58 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: 0.62 AUP1ancient ubiquitous protein 1 (220525_s_at), score: 0.52 B4GALT5UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 (221485_at), score: 0.57 BAXBCL2-associated X protein (211833_s_at), score: -0.57 BAZ2Bbromodomain adjacent to zinc finger domain, 2B (203080_s_at), score: -0.67 BCAP31B-cell receptor-associated protein 31 (200837_at), score: 0.5 BHLHB9basic helix-loop-helix domain containing, class B, 9 (213709_at), score: -0.69 BRF2BRF2, subunit of RNA polymerase III transcription initiation factor, BRF1-like (218955_at), score: 0.64 BTN3A1butyrophilin, subfamily 3, member A1 (209770_at), score: -0.65 C12orf35chromosome 12 open reading frame 35 (218614_at), score: 0.52 C12orf47chromosome 12 open reading frame 47 (64432_at), score: 0.55 C14orf169chromosome 14 open reading frame 169 (219526_at), score: -0.74 C15orf29chromosome 15 open reading frame 29 (218791_s_at), score: -0.63 C17orf86chromosome 17 open reading frame 86 (221621_at), score: 0.51 C20orf11chromosome 20 open reading frame 11 (218448_at), score: 0.65 C20orf111chromosome 20 open reading frame 111 (209020_at), score: 0.7 C20orf4chromosome 20 open reading frame 4 (218089_at), score: 0.52 C21orf66chromosome 21 open reading frame 66 (221158_at), score: 0.57 C4orf30chromosome 4 open reading frame 30 (219717_at), score: 0.52 C6orf62chromosome 6 open reading frame 62 (222309_at), score: 0.62 C7orf23chromosome 7 open reading frame 23 (204215_at), score: 0.52 C7orf28Bchromosome 7 open reading frame 28B (201973_s_at), score: 0.54 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: 0.55 C8orf33chromosome 8 open reading frame 33 (218187_s_at), score: 0.57 C8orf4chromosome 8 open reading frame 4 (218541_s_at), score: -0.63 C9orf125chromosome 9 open reading frame 125 (213386_at), score: -0.58 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (218168_s_at), score: 0.58 CALM1calmodulin 1 (phosphorylase kinase, delta) (209563_x_at), score: -0.59 CAMSAP1L1calmodulin regulated spectrin-associated protein 1-like 1 (212763_at), score: 0.51 CAND1cullin-associated and neddylation-dissociated 1 (208839_s_at), score: -0.67 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: -0.62 CAPNS1calpain, small subunit 1 (200001_at), score: -0.57 CARD8caspase recruitment domain family, member 8 (204950_at), score: -0.57 CASKcalcium/calmodulin-dependent serine protein kinase (MAGUK family) (211208_s_at), score: -0.66 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (207625_s_at), score: 0.69 CCBL2cysteine conjugate-beta lyase 2 (209472_at), score: -0.61 CCNL2cyclin L2 (221427_s_at), score: 0.52 CD44CD44 molecule (Indian blood group) (210916_s_at), score: 0.63 CD63CD63 molecule (200663_at), score: 0.56 CDK5RAP1CDK5 regulatory subunit associated protein 1 (218315_s_at), score: 0.63 CELSR3cadherin, EGF LAG seven-pass G-type receptor 3 (flamingo homolog, Drosophila) (40020_at), score: 0.79 CHCHD2coiled-coil-helix-coiled-coil-helix domain containing 2 (217720_at), score: 0.53 CHKAcholine kinase alpha (204233_s_at), score: 0.7 CHMP1Achromatin modifying protein 1A (201933_at), score: 0.52 CHMP2Bchromatin modifying protein 2B (202537_s_at), score: 0.54 CHN1chimerin (chimaerin) 1 (212624_s_at), score: -0.56 CIRCBF1 interacting corepressor (209571_at), score: 0.62 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.72 CLIC4chloride intracellular channel 4 (201559_s_at), score: -0.57 CLK4CDC-like kinase 4 (210346_s_at), score: 0.54 CLP1CLP1, cleavage and polyadenylation factor I subunit, homolog (S. cerevisiae) (204370_at), score: 0.53 COBLL1COBL-like 1 (203642_s_at), score: -0.6 COL3A1collagen, type III, alpha 1 (215076_s_at), score: -0.65 COL5A2collagen, type V, alpha 2 (221730_at), score: -0.6 COMMD9COMM domain containing 9 (218072_at), score: 0.52 CORO1Ccoronin, actin binding protein, 1C (221676_s_at), score: -0.66 COX10COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (203858_s_at), score: 0.5 CPS1carbamoyl-phosphate synthetase 1, mitochondrial (204920_at), score: -0.57 CREB3L1cAMP responsive element binding protein 3-like 1 (213059_at), score: -0.57 CUGBP2CUG triplet repeat, RNA binding protein 2 (202158_s_at), score: -0.57 CYTH1cytohesin 1 (202880_s_at), score: 0.49 DAPK1death-associated protein kinase 1 (203139_at), score: -0.62 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: 0.55 DDAH1dimethylarginine dimethylaminohydrolase 1 (209094_at), score: -0.72 DEF8differentially expressed in FDCP 8 homolog (mouse) (219646_at), score: 0.5 DGCR2DiGeorge syndrome critical region gene 2 (214198_s_at), score: 0.59 DGCR8DiGeorge syndrome critical region gene 8 (218650_at), score: 0.51 DGKDdiacylglycerol kinase, delta 130kDa (208072_s_at), score: 0.64 DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (212648_at), score: 0.49 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: 0.69 DLX2distal-less homeobox 2 (207147_at), score: 0.55 DNAJC15DnaJ (Hsp40) homolog, subfamily C, member 15 (218435_at), score: -0.66 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: 0.54 DNAJC3DnaJ (Hsp40) homolog, subfamily C, member 3 (208499_s_at), score: 0.53 DNASE1L1deoxyribonuclease I-like 1 (203912_s_at), score: -0.58 DPYDdihydropyrimidine dehydrogenase (204646_at), score: -0.57 DRG1developmentally regulated GTP binding protein 1 (202810_at), score: 0.53 DSEdermatan sulfate epimerase (218854_at), score: -0.56 DSTYKdual serine/threonine and tyrosine protein kinase (214663_at), score: 0.63 EHD2EH-domain containing 2 (221870_at), score: -0.65 EIF1eukaryotic translation initiation factor 1 (212130_x_at), score: 0.65 EIF4ENIF1eukaryotic translation initiation factor 4E nuclear import factor 1 (218626_at), score: 0.57 ELOVL4elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 4 (219532_at), score: -0.6 EML5echinoderm microtubule associated protein like 5 (213063_at), score: -0.58 EMP3epithelial membrane protein 3 (203729_at), score: -0.61 ENOSF1enolase superfamily member 1 (204142_at), score: -0.73 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (209392_at), score: -0.58 EPB41L1erythrocyte membrane protein band 4.1-like 1 (212339_at), score: 0.65 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.55 ERGIC2ERGIC and golgi 2 (218135_at), score: -0.59 ERGIC3ERGIC and golgi 3 (216032_s_at), score: 0.49 ETNK1ethanolamine kinase 1 (219017_at), score: -0.56 EXT1exostoses (multiple) 1 (201995_at), score: -0.6 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.57 FAM105Afamily with sequence similarity 105, member A (219694_at), score: 0.53 FAM134Cfamily with sequence similarity 134, member C (212697_at), score: 0.51 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.82 FAM65Bfamily with sequence similarity 65, member B (209829_at), score: -0.71 FBN2fibrillin 2 (203184_at), score: -0.65 FBXO41F-box protein 41 (44040_at), score: 0.57 FBXO7F-box protein 7 (201178_at), score: 0.55 FBXW2F-box and WD repeat domain containing 2 (218941_at), score: 0.49 FEM1Bfem-1 homolog b (C. elegans) (212367_at), score: -0.57 FLT1fms-related tyrosine kinase 1 (vascular endothelial growth factor/vascular permeability factor receptor) (222033_s_at), score: -0.7 FOXG1forkhead box G1 (206018_at), score: 0.56 FOXK2forkhead box K2 (203064_s_at), score: 0.51 FOXL2forkhead box L2 (220102_at), score: 0.58 FSTL1follistatin-like 1 (208310_s_at), score: 0.53 FUCA1fucosidase, alpha-L- 1, tissue (202838_at), score: 0.57 FZD5frizzled homolog 5 (Drosophila) (221245_s_at), score: 0.64 GAL3ST4galactose-3-O-sulfotransferase 4 (219815_at), score: -0.57 GALCgalactosylceramidase (204417_at), score: -0.64 GALNT3UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) (203397_s_at), score: -0.63 GFRA1GDNF family receptor alpha 1 (205696_s_at), score: -0.69 GIT1G protein-coupled receptor kinase interacting ArfGAP 1 (218030_at), score: 0.63 GJA1gap junction protein, alpha 1, 43kDa (201667_at), score: -0.8 GORASP2golgi reassembly stacking protein 2, 55kDa (208842_s_at), score: 0.57 GPR126G protein-coupled receptor 126 (213094_at), score: -0.59 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.58 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.64 GPX1glutathione peroxidase 1 (200736_s_at), score: -0.62 GREM1gremlin 1, cysteine knot superfamily, homolog (Xenopus laevis) (218469_at), score: -1 GSTK1glutathione S-transferase kappa 1 (217751_at), score: -0.58 GTF2Bgeneral transcription factor IIB (208066_s_at), score: 0.58 GTPBP1GTP binding protein 1 (219357_at), score: 0.51 H1F0H1 histone family, member 0 (208886_at), score: -0.69 HDAC3histone deacetylase 3 (216326_s_at), score: 0.52 HDAC9histone deacetylase 9 (205659_at), score: 0.53 HEATR2HEAT repeat containing 2 (218460_at), score: 0.49 HEXBhexosaminidase B (beta polypeptide) (201944_at), score: 0.51 HEY1hairy/enhancer-of-split related with YRPW motif 1 (44783_s_at), score: 0.57 HIC2hypermethylated in cancer 2 (212964_at), score: 0.58 HIST1H1Chistone cluster 1, H1c (209398_at), score: 0.58 HLA-Bmajor histocompatibility complex, class I, B (211911_x_at), score: 0.6 HLA-Cmajor histocompatibility complex, class I, C (211799_x_at), score: 0.54 HLA-Fmajor histocompatibility complex, class I, F (204806_x_at), score: 0.54 HOXA10homeobox A10 (213150_at), score: -0.63 HSD17B12hydroxysteroid (17-beta) dehydrogenase 12 (217869_at), score: -0.7 HTATIP2HIV-1 Tat interactive protein 2, 30kDa (209448_at), score: -0.56 HYPKHuntingtin interacting protein K (218680_x_at), score: 0.5 IFI30interferon, gamma-inducible protein 30 (201422_at), score: 0.66 IFITM2interferon induced transmembrane protein 2 (1-8D) (201315_x_at), score: -0.62 IFITM3interferon induced transmembrane protein 3 (1-8U) (212203_x_at), score: -0.59 IFT57intraflagellar transport 57 homolog (Chlamydomonas) (218100_s_at), score: 0.5 IGF2BP2insulin-like growth factor 2 mRNA binding protein 2 (218847_at), score: -0.69 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: 0.56 IKIK cytokine, down-regulator of HLA II (200066_at), score: 0.53 IL23Ainterleukin 23, alpha subunit p19 (211796_s_at), score: -0.73 IMAASLC7A5 pseudogene (208118_x_at), score: 0.63 INAinternexin neuronal intermediate filament protein, alpha (204465_s_at), score: -0.6 IPWimprinted in Prader-Willi syndrome (non-protein coding) (221974_at), score: -0.63 IQCKIQ motif containing K (213392_at), score: 0.64 ISL1ISL LIM homeobox 1 (206104_at), score: -0.73 ITGA1integrin, alpha 1 (214660_at), score: -0.57 ITGA5integrin, alpha 5 (fibronectin receptor, alpha polypeptide) (201389_at), score: -0.62 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.66 JAM3junctional adhesion molecule 3 (212813_at), score: -0.8 JMJD1Ajumonji domain containing 1A (212689_s_at), score: 0.61 KCMF1potassium channel modulatory factor 1 (217938_s_at), score: 0.54 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: 0.6 KCTD2potassium channel tetramerisation domain containing 2 (34858_at), score: 0.6 KIAA0174KIAA0174 (200851_s_at), score: 0.52 KIAA1199KIAA1199 (212942_s_at), score: -0.57 KIF3Bkinesin family member 3B (203943_at), score: 0.67 KLHL5kelch-like 5 (Drosophila) (220682_s_at), score: -0.67 KPNA1karyopherin alpha 1 (importin alpha 5) (202055_at), score: -0.58 KYNUkynureninase (L-kynurenine hydrolase) (217388_s_at), score: 0.62 L1CAML1 cell adhesion molecule (204584_at), score: -0.73 LEF1lymphoid enhancer-binding factor 1 (221558_s_at), score: 0.55 LEPROTleptin receptor overlapping transcript (202378_s_at), score: 0.51 LGALS1lectin, galactoside-binding, soluble, 1 (201105_at), score: -0.61 LHFPlipoma HMGIC fusion partner (218656_s_at), score: -0.66 LMO7LIM domain 7 (202674_s_at), score: -0.63 LOC100132214similar to HSPC047 protein (220692_at), score: 0.5 LOC388796hypothetical LOC388796 (65588_at), score: 0.52 LOC440345hypothetical protein LOC440345 (214984_at), score: 0.51 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: 0.59 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.56 LOC51152melanoma antigen (220771_at), score: 0.53 LOC729034similar to puromycin sensitive aminopeptidase (214107_x_at), score: 0.56 LPPR4plasticity related gene 1 (213496_at), score: -0.7 LRRC16Aleucine rich repeat containing 16A (219573_at), score: -0.66 LUMlumican (201744_s_at), score: -0.76 LXNlatexin (218729_at), score: -0.61 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: 0.6 MAFGv-maf musculoaponeurotic fibrosarcoma oncogene homolog G (avian) (204970_s_at), score: 0.6 MAGOHmago-nashi homolog, proliferation-associated (Drosophila) (210092_at), score: 0.51 MAN2A2mannosidase, alpha, class 2A, member 2 (219999_at), score: 0.54 MANSC1MANSC domain containing 1 (220945_x_at), score: -0.63 ME1malic enzyme 1, NADP(+)-dependent, cytosolic (204058_at), score: -0.59 MED4mediator complex subunit 4 (217843_s_at), score: 0.56 MEIS2Meis homeobox 2 (207480_s_at), score: -0.65 METAP1methionyl aminopeptidase 1 (212673_at), score: -0.58 MGAMAX gene associated (212945_s_at), score: -0.61 MLF1myeloid leukemia factor 1 (204784_s_at), score: 0.59 MLLT3myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 (204918_s_at), score: -0.69 MMEmembrane metallo-endopeptidase (203434_s_at), score: -0.61 MMP1matrix metallopeptidase 1 (interstitial collagenase) (204475_at), score: -0.61 MORC4MORC family CW-type zinc finger 4 (219038_at), score: -0.57 MOXD1monooxygenase, DBH-like 1 (209708_at), score: -0.62 MRPS18Cmitochondrial ribosomal protein S18C (220103_s_at), score: -0.69 MT1Fmetallothionein 1F (217165_x_at), score: -0.56 MT1Gmetallothionein 1G (204745_x_at), score: -0.6 MT1Hmetallothionein 1H (206461_x_at), score: -0.58 MT2Ametallothionein 2A (212185_x_at), score: -0.58 MTDHmetadherin (212251_at), score: 0.5 MTERFmitochondrial transcription termination factor (204871_at), score: 0.54 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.85 MUDENGMU-2/AP1M2 domain containing, death-inducing (218139_s_at), score: -0.59 MYO1Bmyosin IB (212364_at), score: -0.81 NAE1NEDD8 activating enzyme E1 subunit 1 (202268_s_at), score: 0.5 NAP1L1nucleosome assembly protein 1-like 1 (208753_s_at), score: -0.68 NBEAneurobeachin (221207_s_at), score: -0.75 NBPF10neuroblastoma breakpoint family, member 10 (214693_x_at), score: 0.57 NCAPH2non-SMC condensin II complex, subunit H2 (40640_at), score: -0.56 NCDNneurochondrin (209556_at), score: -0.58 NCOA6nuclear receptor coactivator 6 (208979_at), score: 0.54 NCRNA00081non-protein coding RNA 81 (213220_at), score: -0.62 NCRNA00115non-protein coding RNA 115 (220399_at), score: 0.6 NDUFAB1NADH dehydrogenase (ubiquinone) 1, alpha/beta subcomplex, 1, 8kDa (202077_at), score: 0.5 NDUFB4NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 4, 15kDa (218226_s_at), score: -0.65 NESnestin (218678_at), score: -0.63 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: -0.6 NGDNneuroguidin, EIF4E binding protein (213794_s_at), score: 0.56 NGFRAP1nerve growth factor receptor (TNFRSF16) associated protein 1 (217963_s_at), score: -0.58 NLGN1neuroligin 1 (205893_at), score: -0.56 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (217796_s_at), score: 0.55 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: 0.49 NRN1neuritin 1 (218625_at), score: 0.67 NRP1neuropilin 1 (212298_at), score: -0.63 NSFL1CNSFL1 (p97) cofactor (p47) (217831_s_at), score: 0.51 NT5E5'-nucleotidase, ecto (CD73) (203939_at), score: -0.63 NUAK2NUAK family, SNF1-like kinase, 2 (220987_s_at), score: -0.62 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: -0.61 NXF1nuclear RNA export factor 1 (208922_s_at), score: 0.52 OSBPL2oxysterol binding protein-like 2 (209222_s_at), score: 0.49 PABPC1poly(A) binding protein, cytoplasmic 1 (215823_x_at), score: -0.56 PABPC3poly(A) binding protein, cytoplasmic 3 (208113_x_at), score: -0.63 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: 0.58 PARP6poly (ADP-ribose) polymerase family, member 6 (219639_x_at), score: 0.65 PBXIP1pre-B-cell leukemia homeobox interacting protein 1 (214176_s_at), score: -0.61 PCGF2polycomb group ring finger 2 (203793_x_at), score: 0.55 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: 0.53 PDCD2programmed cell death 2 (213581_at), score: 0.57 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: -0.75 PDLIM4PDZ and LIM domain 4 (214175_x_at), score: -0.58 PEX1peroxisomal biogenesis factor 1 (204873_at), score: 0.55 PEX19peroxisomal biogenesis factor 19 (201707_at), score: 0.52 PEX5peroxisomal biogenesis factor 5 (203244_at), score: 0.69 PFASphosphoribosylformylglycinamidine synthase (213302_at), score: -0.57 PFTK1PFTAIRE protein kinase 1 (204604_at), score: -0.65 PGAP1post-GPI attachment to proteins 1 (213469_at), score: -0.59 PHF1PHD finger protein 1 (40446_at), score: 0.49 PHF15PHD finger protein 15 (212660_at), score: 0.5 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: 0.6 PHLDA2pleckstrin homology-like domain, family A, member 2 (209803_s_at), score: -0.58 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.52 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.56 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (219048_at), score: -0.72 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.6 PLCB1phospholipase C, beta 1 (phosphoinositide-specific) (213222_at), score: -0.59 PMS2PMS2 postmeiotic segregation increased 2 (S. cerevisiae) (209805_at), score: 0.6 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: 0.77 POFUT1protein O-fucosyltransferase 1 (212349_at), score: 0.66 POLR2Kpolymerase (RNA) II (DNA directed) polypeptide K, 7.0kDa (202634_at), score: 0.49 POM121CPOM121 membrane glycoprotein C (213360_s_at), score: 0.66 PPARGC1Aperoxisome proliferator-activated receptor gamma, coactivator 1 alpha (219195_at), score: 0.64 PPIApeptidylprolyl isomerase A (cyclophilin A) (211378_x_at), score: -0.64 PPIBpeptidylprolyl isomerase B (cyclophilin B) (200968_s_at), score: 0.49 PPIEpeptidylprolyl isomerase E (cyclophilin E) (210502_s_at), score: 0.52 PPP3CBprotein phosphatase 3 (formerly 2B), catalytic subunit, beta isoform (209817_at), score: -0.6 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (202741_at), score: -0.59 PRR16proline rich 16 (220014_at), score: -0.67 PSMB3proteasome (prosome, macropain) subunit, beta type, 3 (201400_at), score: 0.56 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (202352_s_at), score: 0.5 PSMD5proteasome (prosome, macropain) 26S subunit, non-ATPase, 5 (203447_at), score: -0.76 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (219938_s_at), score: -0.68 PTPN1protein tyrosine phosphatase, non-receptor type 1 (202716_at), score: 0.51 PTPN13protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) (204201_s_at), score: -0.57 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.76 PVT1Pvt1 oncogene (non-protein coding) (216249_at), score: 0.53 PYGO1pygopus homolog 1 (Drosophila) (215517_at), score: 0.53 R3HDM2R3H domain containing 2 (203831_at), score: -0.58 RAB22ARAB22A, member RAS oncogene family (218360_at), score: 0.53 RAB2ARAB2A, member RAS oncogene family (208734_x_at), score: 0.52 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: 0.49 RAI14retinoic acid induced 14 (202052_s_at), score: -0.56 RBBP6retinoblastoma binding protein 6 (212781_at), score: 0.49 RECKreversion-inducing-cysteine-rich protein with kazal motifs (205407_at), score: -0.62 REEP5receptor accessory protein 5 (208872_s_at), score: 0.49 RGNEFRho-guanine nucleotide exchange factor (219610_at), score: 0.66 RGS4regulator of G-protein signaling 4 (204337_at), score: -0.59 RHOBras homolog gene family, member B (212099_at), score: -0.59 RIPK4receptor-interacting serine-threonine kinase 4 (221215_s_at), score: 0.64 RLFrearranged L-myc fusion (204243_at), score: 0.5 ROBO1roundabout, axon guidance receptor, homolog 1 (Drosophila) (213194_at), score: -0.75 RP11-138L21.1similar to cell recognition molecule CASPR3 (220436_at), score: 0.51 RP3-377H14.5hypothetical LOC285830 (222279_at), score: 0.51 RPL12ribosomal protein L12 (214271_x_at), score: -0.56 RPL22ribosomal protein L22 (221726_at), score: -0.66 RPL30ribosomal protein L30 (200062_s_at), score: 0.59 RPRD1Aregulation of nuclear pre-mRNA domain containing 1A (218209_s_at), score: -0.58 RPS15ribosomal protein S15 (200819_s_at), score: -0.66 RPS20ribosomal protein S20 (216247_at), score: -0.58 RPS27Lribosomal protein S27-like (218007_s_at), score: -0.56 RPS4P17ribosomal protein S4X pseudogene 17 (216342_x_at), score: 0.49 RPS4Xribosomal protein S4, X-linked (200933_x_at), score: 0.49 RSAD1radical S-adenosyl methionine domain containing 1 (218307_at), score: 0.6 RWDD3RWD domain containing 3 (205087_at), score: 0.51 S100A11S100 calcium binding protein A11 (200660_at), score: -0.76 S100A11PS100 calcium binding protein A11 pseudogene (208540_x_at), score: -0.61 S100A4S100 calcium binding protein A4 (203186_s_at), score: -0.67 SAP18Sin3A-associated protein, 18kDa (208740_at), score: 0.6 SAP30BPSAP30 binding protein (217965_s_at), score: 0.53 SAP30LSAP30-like (219129_s_at), score: 0.57 SAPS2SAPS domain family, member 2 (202792_s_at), score: 0.51 SCAND2SCAN domain containing 2 pseudogene (222177_s_at), score: 0.59 SCPEP1serine carboxypeptidase 1 (218217_at), score: 0.5 SCRIBscribbled homolog (Drosophila) (212556_at), score: -0.61 SDCCAG1serologically defined colon cancer antigen 1 (218649_x_at), score: -0.57 SEH1LSEH1-like (S. cerevisiae) (221931_s_at), score: -0.68 SERP1stress-associated endoplasmic reticulum protein 1 (200971_s_at), score: -0.66 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: -0.65 SETD1ASET domain containing 1A (213202_at), score: 0.66 SGK1serum/glucocorticoid regulated kinase 1 (201739_at), score: -0.59 SIM1single-minded homolog 1 (Drosophila) (206876_at), score: -0.6 SIM2single-minded homolog 2 (Drosophila) (206558_at), score: 0.61 SIRT7sirtuin (silent mating type information regulation 2 homolog) 7 (S. cerevisiae) (218797_s_at), score: 0.5 SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.49 SLC24A1solute carrier family 24 (sodium/potassium/calcium exchanger), member 1 (206081_at), score: -0.67 SLC25A28solute carrier family 25, member 28 (221432_s_at), score: 0.62 SLC25A36solute carrier family 25, member 36 (201918_at), score: 0.57 SLC35F5solute carrier family 35, member F5 (220123_at), score: -0.58 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: -0.68 SLC3A1solute carrier family 3 (cystine, dibasic and neutral amino acid transporters, activator of cystine, dibasic and neutral amino acid transport), member 1 (205799_s_at), score: -0.65 SLC48A1solute carrier family 48 (heme transporter), member 1 (218417_s_at), score: -0.63 SMA4glucuronidase, beta pseudogene (215599_at), score: 0.53 SMA5glucuronidase, beta pseudogene (215043_s_at), score: 0.61 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (221516_s_at), score: 0.57 SNF8SNF8, ESCRT-II complex subunit, homolog (S. cerevisiae) (218391_at), score: 0.56 SNNstannin (218032_at), score: 0.57 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: 0.52 SPATA1spermatogenesis associated 1 (221057_at), score: 0.49 SPCS3signal peptidase complex subunit 3 homolog (S. cerevisiae) (218817_at), score: -0.62 SPHARS-phase response (cyclin-related) (206272_at), score: -0.61 ST6GAL1ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 (201998_at), score: -0.63 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: -0.65 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (204226_at), score: -0.69 STK4serine/threonine kinase 4 (211085_s_at), score: 0.53 STRAPserine/threonine kinase receptor associated protein (200870_at), score: 0.54 STX3syntaxin 3 (209238_at), score: 0.58 STX7syntaxin 7 (212631_at), score: -0.66 SULF1sulfatase 1 (212353_at), score: -0.59 SYNGR1synaptogyrin 1 (210613_s_at), score: -0.72 SYNMsynemin, intermediate filament protein (212730_at), score: -0.67 SYT11synaptotagmin XI (209198_s_at), score: -0.72 TAX1BP1Tax1 (human T-cell leukemia virus type I) binding protein 1 (200976_s_at), score: 0.58 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: -0.63 TBC1D9TBC1 domain family, member 9 (with GRAM domain) (212956_at), score: -0.56 TBXA2Rthromboxane A2 receptor (336_at), score: -0.76 TEStestis derived transcript (3 LIM domains) (202720_at), score: -0.72 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.63 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: 0.5 THAP1THAP domain containing, apoptosis associated protein 1 (219292_at), score: -0.57 THBS2thrombospondin 2 (203083_at), score: -0.72 THSD1thrombospondin, type I, domain containing 1 (219477_s_at), score: 0.61 TMBIM1transmembrane BAX inhibitor motif containing 1 (217730_at), score: 0.61 TMEM111transmembrane protein 111 (217882_at), score: 0.72 TMEM168transmembrane protein 168 (218962_s_at), score: -0.75 TMEM2transmembrane protein 2 (218113_at), score: -0.76 TMEM209transmembrane protein 209 (222267_at), score: 0.53 TMEM223transmembrane protein 223 (220934_s_at), score: -0.57 TMEM45Atransmembrane protein 45A (219410_at), score: -0.62 TMEM87Atransmembrane protein 87A (212202_s_at), score: 0.56 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (210260_s_at), score: -0.57 TNS1tensin 1 (221748_s_at), score: -0.62 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: 0.52 TPBGtrophoblast glycoprotein (203476_at), score: -0.63 TPD52L2tumor protein D52-like 2 (201379_s_at), score: 0.51 TRHDEthyrotropin-releasing hormone degrading enzyme (219937_at), score: -0.65 TRIM32tripartite motif-containing 32 (203846_at), score: -0.68 TRIM52tripartite motif-containing 52 (221897_at), score: 0.52 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: 0.61 TRNAG6transfer RNA glycine 6 (anticodon GCC) (217542_at), score: -0.67 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: -0.56 TSPAN6tetraspanin 6 (209108_at), score: -0.61 TSPYL5TSPY-like 5 (213122_at), score: -0.63 TTC19tetratricopeptide repeat domain 19 (217964_at), score: 0.6 TTC27tetratricopeptide repeat domain 27 (218710_at), score: -0.58 TUBB6tubulin, beta 6 (209191_at), score: -0.68 UBBubiquitin B (200633_at), score: -0.61 UBE2G2ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) (209042_s_at), score: 0.56 UFD1Lubiquitin fusion degradation 1 like (yeast) (209103_s_at), score: 0.53 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.59 UIMC1ubiquitin interaction motif containing 1 (220746_s_at), score: 0.65 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.58 ULK2unc-51-like kinase 2 (C. elegans) (204062_s_at), score: -0.66 UNC13Bunc-13 homolog B (C. elegans) (202893_at), score: -0.6 UNC84Aunc-84 homolog A (C. elegans) (214169_at), score: 0.69 UPP1uridine phosphorylase 1 (203234_at), score: 0.55 USP7ubiquitin specific peptidase 7 (herpes virus-associated) (201498_at), score: 0.5 VAMP2vesicle-associated membrane protein 2 (synaptobrevin 2) (201557_at), score: 0.68 VGLL3vestigial like 3 (Drosophila) (220327_at), score: -0.59 WACWW domain containing adaptor with coiled-coil (219679_s_at), score: 0.59 WASF1WAS protein family, member 1 (204165_at), score: -0.74 WASLWiskott-Aldrich syndrome-like (205809_s_at), score: 0.62 WBSCR22Williams Beuren syndrome chromosome region 22 (207628_s_at), score: 0.56 WHAMML1WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 (213908_at), score: 0.59 WNT5Awingless-type MMTV integration site family, member 5A (213425_at), score: -0.71 WT1Wilms tumor 1 (206067_s_at), score: 0.55 ZFRzinc finger RNA binding protein (33148_at), score: 0.5 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: -0.58 ZNF140zinc finger protein 140 (204523_at), score: -0.64 ZNF167zinc finger protein 167 (206314_at), score: -0.59 ZNF207zinc finger protein 207 (200828_s_at), score: 0.52 ZNF212zinc finger protein 212 (203985_at), score: 0.69 ZNF215zinc finger protein 215 (220214_at), score: -0.66 ZNF273zinc finger protein 273 (215239_x_at), score: 0.54 ZNF354Azinc finger protein 354A (205427_at), score: 0.63 ZNF415zinc finger protein 415 (205514_at), score: 0.51 ZNF432zinc finger protein 432 (219848_s_at), score: 0.51 ZNF434zinc finger protein 434 (218937_at), score: 0.61 ZNF440zinc finger protein 440 (215892_at), score: 0.54 ZNF444zinc finger protein 444 (50376_at), score: -0.57 ZNF512Bzinc finger protein 512B (55872_at), score: 0.49 ZNF518Azinc finger protein 518A (204291_at), score: -0.61 ZNF639zinc finger protein 639 (218413_s_at), score: 0.49 ZNF652zinc finger protein 652 (205594_at), score: 0.59 ZNF706zinc finger protein 706 (218059_at), score: 0.51 ZSCAN12zinc finger and SCAN domain containing 12 (206507_at), score: 0.49
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-GEOD-4219-raw-cel-1311956824.cel | 24 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956178.cel | 6 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956358.cel | 10 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956418.cel | 13 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956398.cel | 12 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |
| E-GEOD-4219-raw-cel-1311956275.cel | 8 | 7 | Sph-mono | hgu133plus2 | none | Sph-mon 1 |