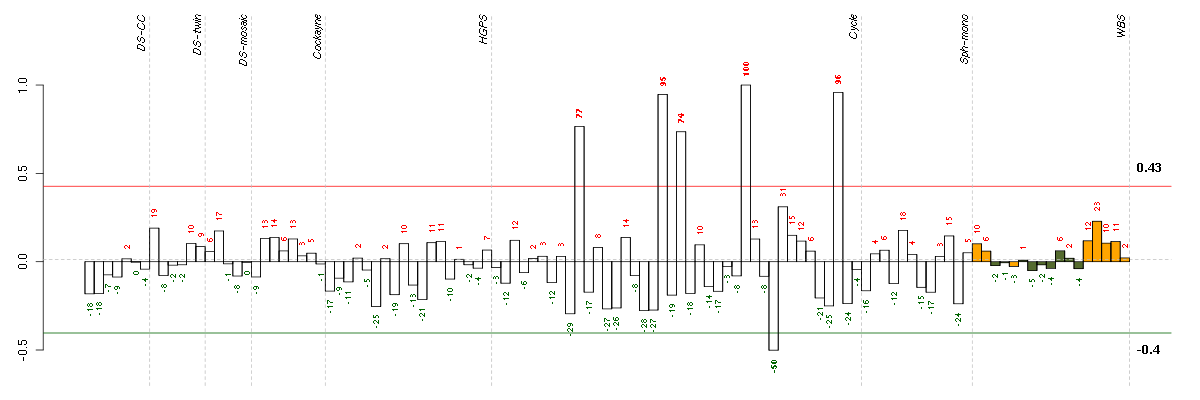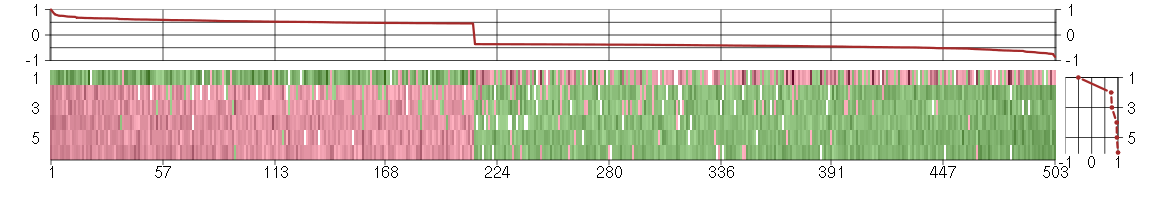

Under-expression is coded with green,
over-expression with red color.

cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures.
organelle organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of a cellular component.
intermediate filament-based process
Any cellular process that depends upon or alters the intermediate filament cytoskeleton, that part of the cytoskeleton comprising intermediate filaments and their associated proteins.
intermediate filament cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising intermediate filaments and their associated proteins.
intermediate filament organization
Control of the spatial distribution of intermediate filaments; includes organizing filaments into meshworks, bundles, or other structures, as by cross-linking.
all
This term is the most general term possible
intermediate filament cytoskeleton organization
A process that is carried out at the cellular level which results in the formation, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising intermediate filaments and their associated proteins.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
all
This term is the most general term possible

AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.46 ACANaggrecan (205679_x_at), score: 0.65 ADAM21ADAM metallopeptidase domain 21 (207665_at), score: 0.47 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: -0.39 ADCK2aarF domain containing kinase 2 (221893_s_at), score: -0.41 ADORA2Badenosine A2b receptor (205891_at), score: 0.46 ADSSadenylosuccinate synthase (221761_at), score: -0.36 AFF2AF4/FMR2 family, member 2 (206105_at), score: -0.61 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.51 AHCTF1AT hook containing transcription factor 1 (214766_s_at), score: -0.37 AHI1Abelson helper integration site 1 (221569_at), score: -0.37 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.48 ANKS1Aankyrin repeat and sterile alpha motif domain containing 1A (212747_at), score: 0.47 ANXA10annexin A10 (210143_at), score: -0.67 AP4S1adaptor-related protein complex 4, sigma 1 subunit (210278_s_at), score: 0.47 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.45 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.54 APOC4apolipoprotein C-IV (206738_at), score: -0.39 ARHGAP8Rho GTPase activating protein 8 (37117_at), score: -0.52 ARMC9armadillo repeat containing 9 (219637_at), score: 0.65 ARNT2aryl-hydrocarbon receptor nuclear translocator 2 (202986_at), score: 0.68 ASB1ankyrin repeat and SOCS box-containing 1 (212819_at), score: -0.42 ASCC1activating signal cointegrator 1 complex subunit 1 (219336_s_at), score: 0.48 ASPNasporin (219087_at), score: -0.45 ASTN2astrotactin 2 (209693_at), score: 0.66 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: 0.57 ATRNL1attractin-like 1 (213744_at), score: -0.36 ATXN3ataxin 3 (205415_s_at), score: 0.48 AXLAXL receptor tyrosine kinase (202685_s_at), score: 0.46 B3GNT2UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 2 (219326_s_at), score: -0.41 B3GNTL1UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase-like 1 (213589_s_at), score: 0.6 BAALCbrain and acute leukemia, cytoplasmic (218899_s_at), score: -0.41 BAHD1bromo adjacent homology domain containing 1 (203051_at), score: -0.39 BAXBCL2-associated X protein (211833_s_at), score: 0.58 BCCIPBRCA2 and CDKN1A interacting protein (218264_at), score: 0.65 BCL11AB-cell CLL/lymphoma 11A (zinc finger protein) (219497_s_at), score: -0.49 BCL2B-cell CLL/lymphoma 2 (203685_at), score: -0.36 BCS1LBCS1-like (yeast) (207618_s_at), score: 0.5 BIRC3baculoviral IAP repeat-containing 3 (210538_s_at), score: -0.38 BTBD3BTB (POZ) domain containing 3 (202946_s_at), score: -0.44 C10orf88chromosome 10 open reading frame 88 (219240_s_at), score: -0.38 C11orf51chromosome 11 open reading frame 51 (204218_at), score: 0.54 C11orf67chromosome 11 open reading frame 67 (221600_s_at), score: 0.65 C11orf71chromosome 11 open reading frame 71 (218789_s_at), score: 0.46 C12orf5chromosome 12 open reading frame 5 (219099_at), score: 0.47 C12orf52chromosome 12 open reading frame 52 (221777_at), score: 0.48 C13orf27chromosome 13 open reading frame 27 (213346_at), score: -0.4 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.38 C16orf62chromosome 16 open reading frame 62 (203173_s_at), score: 0.48 C17orf39chromosome 17 open reading frame 39 (220058_at), score: 0.65 C19orf22chromosome 19 open reading frame 22 (221764_at), score: -0.4 C19orf54chromosome 19 open reading frame 54 (222052_at), score: 0.61 C1orf115chromosome 1 open reading frame 115 (218546_at), score: -0.38 C1orf174chromosome 1 open reading frame 174 (222000_at), score: -0.38 C1orf50chromosome 1 open reading frame 50 (219406_at), score: 0.5 C1orf66chromosome 1 open reading frame 66 (218914_at), score: 0.45 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.61 C21orf7chromosome 21 open reading frame 7 (221211_s_at), score: -0.43 C21orf91chromosome 21 open reading frame 91 (220941_s_at), score: 0.62 C22orf29chromosome 22 open reading frame 29 (219560_at), score: 0.47 C2CD2LC2CD2-like (204757_s_at), score: -0.62 C2orf49chromosome 2 open reading frame 49 (219662_at), score: 0.47 C5orf23chromosome 5 open reading frame 23 (219054_at), score: -0.41 C8orf41chromosome 8 open reading frame 41 (219124_at), score: 0.51 C9orf114chromosome 9 open reading frame 114 (218565_at), score: 0.58 CA1carbonic anhydrase I (205949_at), score: -0.43 CA12carbonic anhydrase XII (203963_at), score: 0.59 CALB1calbindin 1, 28kDa (205626_s_at), score: -0.36 CAND2cullin-associated and neddylation-dissociated 2 (putative) (213547_at), score: 0.46 CAPZA1capping protein (actin filament) muscle Z-line, alpha 1 (208374_s_at), score: 0.46 CASD1CAS1 domain containing 1 (219342_at), score: 0.52 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.48 CCDC28Bcoiled-coil domain containing 28B (221912_s_at), score: 0.46 CCDC33coiled-coil domain containing 33 (220908_at), score: -0.41 CCDC44coiled-coil domain containing 44 (221069_s_at), score: 0.58 CCDC94coiled-coil domain containing 94 (204335_at), score: 0.46 CCL11chemokine (C-C motif) ligand 11 (210133_at), score: -0.5 CCNE1cyclin E1 (213523_at), score: -0.36 CCNT2cyclin T2 (204645_at), score: -0.41 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: -0.38 CD226CD226 molecule (207315_at), score: -0.37 CD320CD320 molecule (218529_at), score: 0.66 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.6 CD83CD83 molecule (204440_at), score: -0.41 CDC25Acell division cycle 25 homolog A (S. pombe) (204695_at), score: -0.45 CDCA4cell division cycle associated 4 (218399_s_at), score: -0.48 CDCP1CUB domain containing protein 1 (218451_at), score: -0.6 CDH18cadherin 18, type 2 (206280_at), score: 0.58 CDKL3cyclin-dependent kinase-like 3 (219831_at), score: 0.65 CDT1chromatin licensing and DNA replication factor 1 (209832_s_at), score: -0.39 CENPTcentromere protein T (218148_at), score: -0.45 CEP70centrosomal protein 70kDa (219036_at), score: 0.53 CHD2chromodomain helicase DNA binding protein 2 (203461_at), score: -0.39 CHRM2cholinergic receptor, muscarinic 2 (221330_at), score: 0.53 CHRNA6cholinergic receptor, nicotinic, alpha 6 (207568_at), score: -0.36 CLCA2chloride channel accessory 2 (217528_at), score: -0.36 CLCF1cardiotrophin-like cytokine factor 1 (219500_at), score: -0.45 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: 0.47 CLEC1AC-type lectin domain family 1, member A (219761_at), score: -0.4 CLEC2DC-type lectin domain family 2, member D (220132_s_at), score: -0.52 CLEC3BC-type lectin domain family 3, member B (205200_at), score: -0.72 CLK1CDC-like kinase 1 (214683_s_at), score: -0.48 CLK4CDC-like kinase 4 (210346_s_at), score: -0.5 CLUclusterin (208791_at), score: -0.53 CNKSR2connector enhancer of kinase suppressor of Ras 2 (206731_at), score: -0.55 CNNM2cyclin M2 (206818_s_at), score: 0.62 CNPY3canopy 3 homolog (zebrafish) (217931_at), score: 0.5 COG2component of oligomeric golgi complex 2 (203073_at), score: 0.49 COL14A1collagen, type XIV, alpha 1 (212865_s_at), score: -0.52 COL5A3collagen, type V, alpha 3 (52255_s_at), score: -0.74 COL7A1collagen, type VII, alpha 1 (217312_s_at), score: -0.49 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: 0.59 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (213379_at), score: 0.51 CORO2Bcoronin, actin binding protein, 2B (209789_at), score: -0.66 COX15COX15 homolog, cytochrome c oxidase assembly protein (yeast) (221550_at), score: 0.58 CPA4carboxypeptidase A4 (205832_at), score: 0.49 CPT1Acarnitine palmitoyltransferase 1A (liver) (203633_at), score: 0.55 CRYL1crystallin, lambda 1 (220753_s_at), score: -0.43 CST6cystatin E/M (206595_at), score: -0.62 CTSEcathepsin E (205927_s_at), score: -0.42 CTSKcathepsin K (202450_s_at), score: -0.52 CTSOcathepsin O (203758_at), score: -0.56 CTSScathepsin S (202901_x_at), score: -0.53 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: -0.48 CXorf15chromosome X open reading frame 15 (219969_at), score: -0.48 CYBBcytochrome b-245, beta polypeptide (203923_s_at), score: -0.38 CYorf14chromosome Y open reading frame 14 (207063_at), score: -0.37 CYP11B1cytochrome P450, family 11, subfamily B, polypeptide 1 (214610_at), score: -0.39 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (211440_x_at), score: -0.4 DARS2aspartyl-tRNA synthetase 2, mitochondrial (218365_s_at), score: 0.54 DCKdeoxycytidine kinase (203302_at), score: -0.39 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.46 DCPSdecapping enzyme, scavenger (218774_at), score: 0.52 DDR1discoidin domain receptor tyrosine kinase 1 (210749_x_at), score: 0.53 DDX43DEAD (Asp-Glu-Ala-Asp) box polypeptide 43 (220004_at), score: 0.53 DDX49DEAD (Asp-Glu-Ala-Asp) box polypeptide 49 (31807_at), score: 0.46 DEPDC6DEP domain containing 6 (218858_at), score: 0.47 DHRS2dehydrogenase/reductase (SDR family) member 2 (214079_at), score: -0.44 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.47 DNAJC7DnaJ (Hsp40) homolog, subfamily C, member 7 (202416_at), score: 0.47 DNAJC8DnaJ (Hsp40) homolog, subfamily C, member 8 (205545_x_at), score: 0.48 DNM3dynamin 3 (209839_at), score: -0.43 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: -0.55 DSCR3Down syndrome critical region gene 3 (203635_at), score: 0.61 DSN1DSN1, MIND kinetochore complex component, homolog (S. cerevisiae) (219512_at), score: -0.42 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (205761_s_at), score: 0.59 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.37 DUSP5dual specificity phosphatase 5 (209457_at), score: -0.52 DUSP6dual specificity phosphatase 6 (208892_s_at), score: -0.46 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.49 EDNRBendothelin receptor type B (204271_s_at), score: -0.38 EGLN3egl nine homolog 3 (C. elegans) (219232_s_at), score: -0.38 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.36 EIF3Heukaryotic translation initiation factor 3, subunit H (201592_at), score: 0.49 EIF4EBP2eukaryotic translation initiation factor 4E binding protein 2 (208770_s_at), score: -0.43 ELA3Aelastase 3A, pancreatic (211738_x_at), score: -0.48 ELK3ELK3, ETS-domain protein (SRF accessory protein 2) (221773_at), score: -0.36 ELK4ELK4, ETS-domain protein (SRF accessory protein 1) (206919_at), score: -0.37 EMCNendomucin (219436_s_at), score: 0.67 EML1echinoderm microtubule associated protein like 1 (204797_s_at), score: -0.57 EREGepiregulin (205767_at), score: -0.49 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.5 ESRRAestrogen-related receptor alpha (1487_at), score: -0.41 EVI2Aecotropic viral integration site 2A (204774_at), score: 0.45 EVI2Becotropic viral integration site 2B (211742_s_at), score: 0.63 EVLEnah/Vasp-like (217838_s_at), score: -0.37 EXOSC5exosome component 5 (218481_at), score: 0.62 FAM117Afamily with sequence similarity 117, member A (221249_s_at), score: 0.54 FAM131Bfamily with sequence similarity 131, member B (205368_at), score: -0.42 FAM13Bfamily with sequence similarity 13, member B (218518_at), score: -0.37 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: -0.44 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.53 FAM63Bfamily with sequence similarity 63, member B (214691_x_at), score: -0.47 FAM70Afamily with sequence similarity 70, member A (219895_at), score: -0.37 FAM86Cfamily with sequence similarity 86, member C (220353_at), score: 0.61 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: -0.36 FBXO31F-box protein 31 (219785_s_at), score: 0.56 FDXRferredoxin reductase (207813_s_at), score: 0.53 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.42 FGD2FYVE, RhoGEF and PH domain containing 2 (215602_at), score: -0.54 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: -0.38 FURINfurin (paired basic amino acid cleaving enzyme) (201945_at), score: -0.37 FUT4fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) (209892_at), score: -0.38 FUZfuzzy homolog (Drosophila) (221187_s_at), score: 0.45 FZD4frizzled homolog 4 (Drosophila) (218665_at), score: 0.62 GAAglucosidase, alpha; acid (202812_at), score: -0.55 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.44 GALTgalactose-1-phosphate uridylyltransferase (203179_at), score: 0.59 GANgigaxonin (220124_at), score: -0.43 GBAPglucosidase, beta; acid, pseudogene (210589_s_at), score: 0.48 GDF9growth differentiation factor 9 (221314_at), score: -0.42 GKglycerol kinase (207387_s_at), score: -0.48 GNB1Lguanine nucleotide binding protein (G protein), beta polypeptide 1-like (220762_s_at), score: 0.52 GNPDA1glucosamine-6-phosphate deaminase 1 (202382_s_at), score: 0.54 GPM6Bglycoprotein M6B (209170_s_at), score: -0.71 GPR153G protein-coupled receptor 153 (221902_at), score: -0.45 GPR157G protein-coupled receptor 157 (220901_at), score: -0.36 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.61 GPR183G protein-coupled receptor 183 (205419_at), score: -0.38 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.8 GPSN2glycoprotein, synaptic 2 (208336_s_at), score: 0.52 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: -0.37 GRB14growth factor receptor-bound protein 14 (206204_at), score: -0.44 GRHL2grainyhead-like 2 (Drosophila) (219388_at), score: -0.45 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.46 GYG2glycogenin 2 (215695_s_at), score: 0.52 H2AFJH2A histone family, member J (220936_s_at), score: -0.47 HEATR6HEAT repeat containing 6 (65493_at), score: 0.52 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (209960_at), score: -0.56 HINFPhistone H4 transcription factor (206495_s_at), score: 0.47 HIPK3homeodomain interacting protein kinase 3 (210148_at), score: -0.36 HISPPD2Ahistidine acid phosphatase domain containing 2A (204578_at), score: 0.49 HIST1H2AGhistone cluster 1, H2ag (207156_at), score: 0.58 HIST1H2BIhistone cluster 1, H2bi (208523_x_at), score: -0.37 HIST1H2BKhistone cluster 1, H2bk (209806_at), score: -0.37 HIST1H4Ghistone cluster 1, H4g (208551_at), score: -0.36 HIST1H4Hhistone cluster 1, H4h (208180_s_at), score: -0.39 HOMER2homer homolog 2 (Drosophila) (217080_s_at), score: -0.37 HOXD1homeobox D1 (205975_s_at), score: -0.4 HSD11B1hydroxysteroid (11-beta) dehydrogenase 1 (205404_at), score: -0.36 HSPB2heat shock 27kDa protein 2 (205824_at), score: 0.65 HSPBAP1HSPB (heat shock 27kDa) associated protein 1 (219284_at), score: 0.64 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (207135_at), score: -0.42 IER2immediate early response 2 (202081_at), score: -0.4 IFNGinterferon, gamma (210354_at), score: -0.42 IGF2insulin-like growth factor 2 (somatomedin A) (202409_at), score: -0.52 IL12Ainterleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) (207160_at), score: -0.39 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.62 IL16interleukin 16 (lymphocyte chemoattractant factor) (209827_s_at), score: -0.45 IL33interleukin 33 (209821_at), score: -0.86 INTS8integrator complex subunit 8 (218905_at), score: 0.46 ISYNA1inositol-3-phosphate synthase 1 (222240_s_at), score: 0.49 JMJD1Ajumonji domain containing 1A (212689_s_at), score: 0.46 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.58 KCND2potassium voltage-gated channel, Shal-related subfamily, member 2 (207103_at), score: -0.37 KCNE4potassium voltage-gated channel, Isk-related family, member 4 (222379_at), score: 0.66 KCNJ15potassium inwardly-rectifying channel, subfamily J, member 15 (210119_at), score: -0.37 KCTD13potassium channel tetramerisation domain containing 13 (45653_at), score: -0.38 KCTD9potassium channel tetramerisation domain containing 9 (218823_s_at), score: -0.53 KIAA0146KIAA0146 (212523_s_at), score: -0.36 KIAA0226KIAA0226 (212733_at), score: -0.41 KIAA0415KIAA0415 (209912_s_at), score: 0.58 KIAA0564KIAA0564 (212946_at), score: 0.48 KIAA0586KIAA0586 (205631_at), score: 0.56 KIAA0907KIAA0907 (202220_at), score: -0.46 KIAA1024KIAA1024 (215081_at), score: -0.49 KITv-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog (205051_s_at), score: 0.61 KRT14keratin 14 (209351_at), score: -0.44 KRT17keratin 17 (212236_x_at), score: -0.4 KRT33Akeratin 33A (208483_x_at), score: -0.67 KRT9keratin 9 (208188_at), score: -0.37 LANCL2LanC lantibiotic synthetase component C-like 2 (bacterial) (218219_s_at), score: 0.47 LDB1LIM domain binding 1 (203451_at), score: 0.47 LEMD3LEM domain containing 3 (218604_at), score: -0.42 LIFleukemia inhibitory factor (cholinergic differentiation factor) (205266_at), score: -0.37 LIG3ligase III, DNA, ATP-dependent (204123_at), score: 0.55 LMAN2Llectin, mannose-binding 2-like (221274_s_at), score: 0.68 LOC100128223hypothetical protein LOC100128223 (221264_s_at), score: 0.47 LOC100133558similar to hCG1642170 (216315_x_at), score: -0.39 LOC388152hypothetical LOC388152 (220602_s_at), score: 0.73 LOC390940similar to R28379_1 (213556_at), score: 0.48 LOC65998hypothetical protein LOC65998 (218641_at), score: -0.48 LOC729806similar to hCG1725380 (217544_at), score: -0.41 LPAR4lysophosphatidic acid receptor 4 (206960_at), score: -0.36 LRP2low density lipoprotein-related protein 2 (205710_at), score: -0.36 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.48 MAFFv-maf musculoaponeurotic fibrosarcoma oncogene homolog F (avian) (36711_at), score: -0.37 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: -0.44 MAP4K4mitogen-activated protein kinase kinase kinase kinase 4 (218181_s_at), score: -0.49 MAP7D3MAP7 domain containing 3 (219576_at), score: 0.47 MAPK13mitogen-activated protein kinase 13 (210059_s_at), score: 0.61 MARCH3membrane-associated ring finger (C3HC4) 3 (213256_at), score: -0.36 MAST2microtubule associated serine/threonine kinase 2 (211593_s_at), score: -0.37 MBmyoglobin (204179_at), score: -0.39 MBD5methyl-CpG binding domain protein 5 (220195_at), score: 0.51 MCF2L2MCF.2 cell line derived transforming sequence-like 2 (215112_x_at), score: -0.43 MID1IP1MID1 interacting protein 1 (gastrulation specific G12 homolog (zebrafish)) (218251_at), score: -0.5 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.4 MLPHmelanophilin (218211_s_at), score: 0.67 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.47 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.47 MOSPD1motile sperm domain containing 1 (218853_s_at), score: 0.57 MPImannose phosphate isomerase (202472_at), score: 0.52 MSTP9macrophage stimulating, pseudogene 9 (213382_at), score: -0.66 MTERFmitochondrial transcription termination factor (204871_at), score: 0.51 MTMR1myotubularin related protein 1 (214975_s_at), score: -0.46 MTMR9myotubularin related protein 9 (204837_at), score: -0.38 MUC3Bmucin 3B, cell surface associated (214898_x_at), score: 0.54 MUSKmuscle, skeletal, receptor tyrosine kinase (207633_s_at), score: -0.42 MXD4MAX dimerization protein 4 (210778_s_at), score: 0.47 MYCL1v-myc myelocytomatosis viral oncogene homolog 1, lung carcinoma derived (avian) (214058_at), score: -0.41 N6AMT1N-6 adenine-specific DNA methyltransferase 1 (putative) (220311_at), score: 0.51 NAGPAN-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase (205090_s_at), score: 0.61 NAPGN-ethylmaleimide-sensitive factor attachment protein, gamma (210048_at), score: 0.48 NARS2asparaginyl-tRNA synthetase 2, mitochondrial (putative) (219217_at), score: 0.49 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.4 NBL1neuroblastoma, suppression of tumorigenicity 1 (37005_at), score: -0.4 NCALDneurocalcin delta (211685_s_at), score: -0.37 NDRG4NDRG family member 4 (209159_s_at), score: 0.54 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.46 NEFLneurofilament, light polypeptide (221805_at), score: -0.67 NEFMneurofilament, medium polypeptide (205113_at), score: -0.4 NELFnasal embryonic LHRH factor (221214_s_at), score: -0.37 NFATC2IPnuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 2 interacting protein (217526_at), score: -0.7 NINninein (GSK3B interacting protein) (219285_s_at), score: -0.38 NMBneuromedin B (205204_at), score: 0.49 NMNAT2nicotinamide nucleotide adenylyltransferase 2 (209755_at), score: -0.37 NOL8nucleolar protein 8 (218244_at), score: 0.71 NPnucleoside phosphorylase (201695_s_at), score: -0.43 NR0B1nuclear receptor subfamily 0, group B, member 1 (206644_at), score: -0.61 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.47 NSD1nuclear receptor binding SET domain protein 1 (219084_at), score: 0.53 NTHL1nth endonuclease III-like 1 (E. coli) (209731_at), score: 0.66 NTMneurotrimin (222020_s_at), score: -0.39 NTNG1netrin G1 (206713_at), score: -0.37 NUAK1NUAK family, SNF1-like kinase, 1 (204589_at), score: 0.51 NUDT1nudix (nucleoside diphosphate linked moiety X)-type motif 1 (204766_s_at), score: 0.56 NUDT6nudix (nucleoside diphosphate linked moiety X)-type motif 6 (220183_s_at), score: 0.58 NUPL1nucleoporin like 1 (204435_at), score: -0.49 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: -0.38 OLFML2Aolfactomedin-like 2A (213075_at), score: -0.57 OMDosteomodulin (205907_s_at), score: -0.43 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.47 OR2W1olfactory receptor, family 2, subfamily W, member 1 (221451_s_at), score: -0.36 PAFAH1B3platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa (203228_at), score: 0.64 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: 0.53 PDCphosducin (211496_s_at), score: -0.53 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.55 PEG3paternally expressed 3 (209242_at), score: -0.51 PENKproenkephalin (213791_at), score: -0.49 PET112LPET112-like (yeast) (204300_at), score: 0.6 PF4V1platelet factor 4 variant 1 (207815_at), score: -0.38 PFDN6prefoldin subunit 6 (222029_x_at), score: 0.56 PFTK1PFTAIRE protein kinase 1 (204604_at), score: -0.37 PHKA1phosphorylase kinase, alpha 1 (muscle) (205450_at), score: -0.48 PHLPPPH domain and leucine rich repeat protein phosphatase (212719_at), score: -0.45 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: -0.37 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.58 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.51 PIONpigeon homolog (Drosophila) (222150_s_at), score: -0.39 PKIAprotein kinase (cAMP-dependent, catalytic) inhibitor alpha (204612_at), score: -0.59 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.51 PLATplasminogen activator, tissue (201860_s_at), score: -0.39 PLAURplasminogen activator, urokinase receptor (211924_s_at), score: -0.48 PLCXD1phosphatidylinositol-specific phospholipase C, X domain containing 1 (218951_s_at), score: -0.37 PMAIP1phorbol-12-myristate-13-acetate-induced protein 1 (204286_s_at), score: -0.7 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.38 PNRC2proline-rich nuclear receptor coactivator 2 (217779_s_at), score: -0.65 POP1processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) (213449_at), score: 0.56 POU4F1POU class 4 homeobox 1 (206940_s_at), score: -0.46 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: -0.42 PPM1Aprotein phosphatase 1A (formerly 2C), magnesium-dependent, alpha isoform (210407_at), score: 0.45 PRELPproline/arginine-rich end leucine-rich repeat protein (204223_at), score: -0.45 PRKG2protein kinase, cGMP-dependent, type II (207505_at), score: -0.62 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: -0.61 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.74 PRSS3protease, serine, 3 (207463_x_at), score: -0.75 PSMB9proteasome (prosome, macropain) subunit, beta type, 9 (large multifunctional peptidase 2) (204279_at), score: 0.62 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: 0.61 PTENP1phosphatase and tensin homolog pseudogene 1 (217492_s_at), score: 0.52 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.58 PTGS2prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (204748_at), score: -0.42 PTPRBprotein tyrosine phosphatase, receptor type, B (205846_at), score: -0.42 PTPRDprotein tyrosine phosphatase, receptor type, D (214043_at), score: -0.38 PYCARDPYD and CARD domain containing (221666_s_at), score: 0.55 R3HDM1R3H domain containing 1 (202754_at), score: 0.49 RAB13RAB13, member RAS oncogene family (202252_at), score: 0.58 RAB6CRAB6C, member RAS oncogene family (210406_s_at), score: -0.36 RAB8BRAB8B, member RAS oncogene family (219210_s_at), score: 0.71 RAD54L2RAD54-like 2 (S. cerevisiae) (213205_s_at), score: -0.4 RAGErenal tumor antigen (205130_at), score: -0.43 RALGDSral guanine nucleotide dissociation stimulator (209050_s_at), score: 0.47 RAPGEF4Rap guanine nucleotide exchange factor (GEF) 4 (205651_x_at), score: -0.44 RARRES1retinoic acid receptor responder (tazarotene induced) 1 (221872_at), score: -0.49 RASA2RAS p21 protein activator 2 (206636_at), score: -0.52 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: -0.52 RBL1retinoblastoma-like 1 (p107) (205296_at), score: -0.36 RBM3RNA binding motif (RNP1, RRM) protein 3 (208319_s_at), score: -0.71 RBM39RNA binding motif protein 39 (207941_s_at), score: -0.49 RBM4BRNA binding motif protein 4B (209497_s_at), score: 0.66 REEP1receptor accessory protein 1 (204364_s_at), score: -0.38 RFX7regulatory factor X, 7 (218430_s_at), score: 0.54 RNASE4ribonuclease, RNase A family, 4 (205158_at), score: -0.36 RNF138ring finger protein 138 (218738_s_at), score: -0.37 RNF44ring finger protein 44 (203286_at), score: 0.53 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.56 RPGRIP1retinitis pigmentosa GTPase regulator interacting protein 1 (206608_s_at), score: -0.45 RPL23ribosomal protein L23 (214744_s_at), score: -0.37 RPL27Aribosomal protein L27a (212044_s_at), score: 0.48 RPS11ribosomal protein S11 (213350_at), score: 0.47 RWDD2BRWD domain containing 2B (218377_s_at), score: 0.47 S100A3S100 calcium binding protein A3 (206027_at), score: 0.51 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.38 SARM1sterile alpha and TIR motif containing 1 (213259_s_at), score: 0.51 SAT1spermidine/spermine N1-acetyltransferase 1 (213988_s_at), score: -0.62 SEMA3Csema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C (203788_s_at), score: -0.57 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.4 SERPINB7serpin peptidase inhibitor, clade B (ovalbumin), member 7 (206421_s_at), score: 0.52 SERPING1serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 (200986_at), score: -0.38 SESN1sestrin 1 (218346_s_at), score: 0.47 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.75 SEZ6L2seizure related 6 homolog (mouse)-like 2 (218720_x_at), score: -0.39 SFRS12splicing factor, arginine/serine-rich 12 (212721_at), score: -0.52 SFRS6splicing factor, arginine/serine-rich 6 (206108_s_at), score: 0.49 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.4 SIPA1signal-induced proliferation-associated 1 (204164_at), score: -0.38 SIRPB1signal-regulatory protein beta 1 (206934_at), score: -0.46 SLC12A8solute carrier family 12 (potassium/chloride transporters), member 8 (219874_at), score: 0.48 SLC16A5solute carrier family 16, member 5 (monocarboxylic acid transporter 6) (206600_s_at), score: 0.78 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: -0.47 SLC17A6solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 6 (220551_at), score: -0.49 SLC17A7solute carrier family 17 (sodium-dependent inorganic phosphate cotransporter), member 7 (204230_s_at), score: -0.46 SLC20A2solute carrier family 20 (phosphate transporter), member 2 (202744_at), score: -0.42 SLC22A4solute carrier family 22 (organic cation/ergothioneine transporter), member 4 (205896_at), score: -0.37 SLC26A4solute carrier family 26, member 4 (206529_x_at), score: -0.48 SLC28A3solute carrier family 28 (sodium-coupled nucleoside transporter), member 3 (220475_at), score: -0.43 SLC38A4solute carrier family 38, member 4 (220786_s_at), score: 0.89 SLC44A1solute carrier family 44, member 1 (222364_at), score: 0.54 SLKSTE20-like kinase (yeast) (206875_s_at), score: -0.43 SMU1smu-1 suppressor of mec-8 and unc-52 homolog (C. elegans) (218393_s_at), score: 0.57 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: 0.51 SNX13sorting nexin 13 (213292_s_at), score: 0.55 SORT1sortilin 1 (212807_s_at), score: 0.54 SOX9SRY (sex determining region Y)-box 9 (202935_s_at), score: -0.46 SP1Sp1 transcription factor (214732_at), score: -0.36 SPA17sperm autoantigenic protein 17 (205406_s_at), score: 0.58 SPAG8sperm associated antigen 8 (206816_s_at), score: 0.46 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: -0.38 SPP1secreted phosphoprotein 1 (209875_s_at), score: 0.52 SPRY2sprouty homolog 2 (Drosophila) (204011_at), score: -0.38 SPRY4sprouty homolog 4 (Drosophila) (221489_s_at), score: -0.47 SSFA2sperm specific antigen 2 (202506_at), score: -0.41 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.6 STC1stanniocalcin 1 (204597_x_at), score: -0.41 STX3syntaxin 3 (209238_at), score: 0.46 SUOXsulfite oxidase (204067_at), score: 0.63 SYNMsynemin, intermediate filament protein (212730_at), score: 0.75 SYT1synaptotagmin I (203999_at), score: -0.51 SYT11synaptotagmin XI (209198_s_at), score: 0.75 TBCEtubulin folding cofactor E (203714_s_at), score: 0.46 TBX5T-box 5 (207155_at), score: 0.52 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.72 TGFAtransforming growth factor, alpha (205016_at), score: -0.5 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (205210_at), score: 0.57 THEM2thioesterase superfamily member 2 (204565_at), score: 0.56 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: 0.56 THYN1thymocyte nuclear protein 1 (218491_s_at), score: 0.47 TIMM9translocase of inner mitochondrial membrane 9 homolog (yeast) (218316_at), score: 0.5 TMEM158transmembrane protein 158 (213338_at), score: -0.52 TMEM160transmembrane protein 160 (219219_at), score: 0.51 TMEM22transmembrane protein 22 (219569_s_at), score: -0.42 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.46 TMEM9BTMEM9 domain family, member B (218065_s_at), score: -0.37 TMOD1tropomodulin 1 (203661_s_at), score: -0.42 TMSB15Athymosin beta 15a (205347_s_at), score: 0.74 TMX4thioredoxin-related transmembrane protein 4 (201581_at), score: -0.4 TNIKTRAF2 and NCK interacting kinase (213107_at), score: 0.47 TNS3tensin 3 (217853_at), score: 0.46 TOMM22translocase of outer mitochondrial membrane 22 homolog (yeast) (217960_s_at), score: 0.61 TOXthymocyte selection-associated high mobility group box (204529_s_at), score: 0.58 TPK1thiamin pyrophosphokinase 1 (221218_s_at), score: -0.62 TRAPPC10trafficking protein particle complex 10 (209412_at), score: -0.43 TRIAP1TP53 regulated inhibitor of apoptosis 1 (218403_at), score: 0.6 TRIB2tribbles homolog 2 (Drosophila) (202478_at), score: 0.68 TRPC1transient receptor potential cation channel, subfamily C, member 1 (205802_at), score: -0.53 TRY6trypsinogen C (215395_x_at), score: -0.38 TSPAN13tetraspanin 13 (217979_at), score: -0.37 UAP1UDP-N-acteylglucosamine pyrophosphorylase 1 (209340_at), score: -0.36 UCHL5IPUCHL5 interacting protein (213334_x_at), score: 0.47 UCP2uncoupling protein 2 (mitochondrial, proton carrier) (208998_at), score: 1 USE1unconventional SNARE in the ER 1 homolog (S. cerevisiae) (221706_s_at), score: 0.51 USP47ubiquitin specific peptidase 47 (221518_s_at), score: 0.53 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.72 XDHxanthine dehydrogenase (210301_at), score: -0.36 XPO4exportin 4 (218479_s_at), score: 0.67 YEATS4YEATS domain containing 4 (218911_at), score: -0.43 YIPF1Yip1 domain family, member 1 (214733_s_at), score: 0.46 YY2YY2 transcription factor (216531_at), score: 0.6 ZDHHC18zinc finger, DHHC-type containing 18 (212860_at), score: -0.48 ZFYVE9zinc finger, FYVE domain containing 9 (204893_s_at), score: 0.53 ZHX2zinc fingers and homeoboxes 2 (203556_at), score: 0.5 ZNF174zinc finger protein 174 (210290_at), score: 0.48 ZNF185zinc finger protein 185 (LIM domain) (203585_at), score: -0.37 ZNF195zinc finger protein 195 (204234_s_at), score: 0.66 ZNF219zinc finger protein 219 (219314_s_at), score: -0.59 ZNF22zinc finger protein 22 (KOX 15) (218006_s_at), score: 0.6 ZNF223zinc finger protein 223 (207128_s_at), score: 0.47 ZNF236zinc finger protein 236 (47571_at), score: -0.37 ZNF287zinc finger protein 287 (216710_x_at), score: -0.51 ZNF3zinc finger protein 3 (212684_at), score: 0.46 ZNF304zinc finger protein 304 (207753_at), score: 0.5 ZNF350zinc finger protein 350 (219266_at), score: 0.52 ZNF43zinc finger protein 43 (206695_x_at), score: -0.36 ZNF434zinc finger protein 434 (218937_at), score: 0.48 ZNF443zinc finger protein 443 (205928_at), score: -0.45 ZNF667zinc finger protein 667 (207120_at), score: 0.59 ZNF675zinc finger protein 675 (217547_x_at), score: -0.45 ZNF688zinc finger protein 688 (213527_s_at), score: 0.54 ZNF749zinc finger protein 749 (215289_at), score: -0.43 ZNF85zinc finger protein 85 (206572_x_at), score: -0.36 ZNF93zinc finger protein 93 (208119_s_at), score: -0.36 ZSCAN16zinc finger and SCAN domain containing 16 (219676_at), score: -0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486251.cel | 31 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486051.cel | 21 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486011.cel | 19 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486391.cel | 38 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486191.cel | 28 | 6 | Cycle | hgu133a2 | none | Cycle 1 |