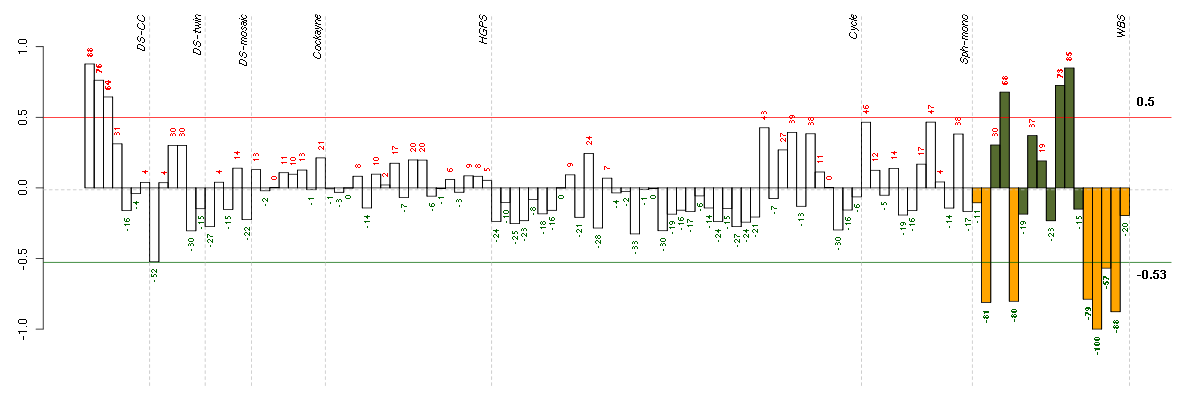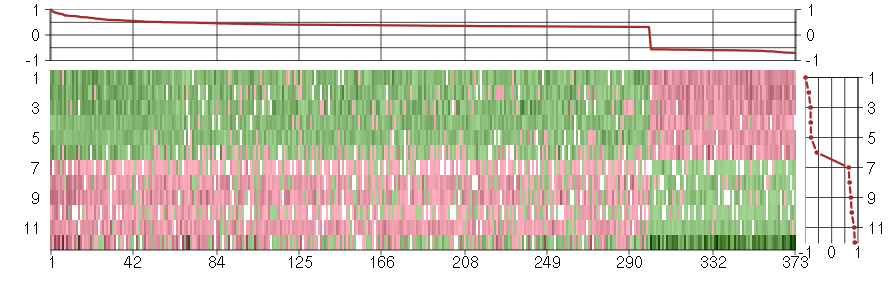

Under-expression is coded with green,
over-expression with red color.

immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to external stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an external stimulus.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
This term is the most general term possible
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
innate immune response
Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens.

extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
proteinaceous extracellular matrix
A layer consisting mainly of proteins (especially collagen) and glycosaminoglycans (mostly as proteoglycans) that forms a sheet underlying or overlying cells such as endothelial and epithelial cells. The proteins are secreted by cells in the vicinity.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
extracellular matrix
A structure lying external to one or more cells, which provides structural support for cells or tissues; may be completely external to the cell (as in animals) or be part of the cell (as in plants).
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
all
This term is the most general term possible
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.

protein binding
Interacting selectively with any protein or protein complex (a complex of two or more proteins that may include other nonprotein molecules).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
cytokine receptor activity
Combining with a cytokine to initiate a change in cell activity.
interleukin-1 receptor activity
Combining with interleukin-1 to initiate a change in cell activity. Interleukin-1 is produced mainly by activated macrophages and is involved in the inflammatory response.
growth factor binding
Interacting selectively with any growth factor, proteins or polypeptides that stimulate a cell or organism to grow or proliferate.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
glutamate receptor activity
Combining with glutamate to initiate a change in cell activity.
cytokine binding
Interacting selectively with a cytokine, any of a group of proteins that function to control the survival, growth and differentiation of tissues and cells, and which have autocrine and paracrine activity.
interleukin-1 binding
Interacting selectively with interleukin-1.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
interleukin-1 binding
Interacting selectively with interleukin-1.
interleukin-1 receptor activity
Combining with interleukin-1 to initiate a change in cell activity. Interleukin-1 is produced mainly by activated macrophages and is involved in the inflammatory response.
cytokine receptor activity
Combining with a cytokine to initiate a change in cell activity.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04060 | 3.262e-02 | 5.191 | 14 | 122 | Cytokine-cytokine receptor interaction |
ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (203504_s_at), score: 0.43 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (213353_at), score: 0.39 ABCC3ATP-binding cassette, sub-family C (CFTR/MRP), member 3 (208161_s_at), score: 0.32 ABCC9ATP-binding cassette, sub-family C (CFTR/MRP), member 9 (208561_at), score: 0.33 ABHD10abhydrolase domain containing 10 (218633_x_at), score: -0.69 ABLIM1actin binding LIM protein 1 (200965_s_at), score: -0.56 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: 0.33 ACVR2Aactivin A receptor, type IIA (205327_s_at), score: 0.64 ADAMTS6ADAM metallopeptidase with thrombospondin type 1 motif, 6 (220866_at), score: 0.39 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.76 ADKadenosine kinase (204119_s_at), score: -0.58 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.32 AKR1A1aldo-keto reductase family 1, member A1 (aldehyde reductase) (201900_s_at), score: -0.63 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.42 ALDH1A3aldehyde dehydrogenase 1 family, member A3 (203180_at), score: 0.64 ALMS1Alstrom syndrome 1 (214707_x_at), score: 0.39 ANKMY2ankyrin repeat and MYND domain containing 2 (212798_s_at), score: -0.6 ANKRD36Bankyrin repeat domain 36B (220940_at), score: 0.35 APODapolipoprotein D (201525_at), score: 0.88 ARHGAP26Rho GTPase activating protein 26 (205068_s_at), score: 0.36 ARHGEF11Rho guanine nucleotide exchange factor (GEF) 11 (202914_s_at), score: 0.32 ARPC1Bactin related protein 2/3 complex, subunit 1B, 41kDa (201954_at), score: -0.6 ATF5activating transcription factor 5 (204999_s_at), score: 0.34 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (220521_s_at), score: 0.4 ATP2B1ATPase, Ca++ transporting, plasma membrane 1 (209281_s_at), score: 0.39 BICC1bicaudal C homolog 1 (Drosophila) (220580_at), score: 0.4 BMP2KBMP2 inducible kinase (219546_at), score: 0.41 BRWD1bromodomain and WD repeat domain containing 1 (214820_at), score: 0.39 BTDbiotinidase (204167_at), score: -0.64 C10orf18chromosome 10 open reading frame 18 (218331_s_at), score: 0.36 C15orf5chromosome 15 open reading frame 5 (208109_s_at), score: 0.7 C17orf91chromosome 17 open reading frame 91 (214696_at), score: 0.39 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.5 C1orf56chromosome 1 open reading frame 56 (221222_s_at), score: 0.42 C1Rcomplement component 1, r subcomponent (212067_s_at), score: 0.4 C1Scomplement component 1, s subcomponent (208747_s_at), score: 0.47 C20orf3chromosome 20 open reading frame 3 (206656_s_at), score: -0.58 C3complement component 3 (217767_at), score: 0.49 C4orf29chromosome 4 open reading frame 29 (219980_at), score: 0.32 C9orf127chromosome 9 open reading frame 127 (207839_s_at), score: -0.61 CAPN3calpain 3, (p94) (210944_s_at), score: 0.39 CCDC76coiled-coil domain containing 76 (219130_at), score: 0.34 CCL7chemokine (C-C motif) ligand 7 (208075_s_at), score: 0.61 CCNL2cyclin L2 (221427_s_at), score: 0.43 CCRL1chemokine (C-C motif) receptor-like 1 (220351_at), score: 0.33 CD226CD226 molecule (207315_at), score: 0.34 CD302CD302 molecule (203799_at), score: 0.33 CDK7cyclin-dependent kinase 7 (211297_s_at), score: -0.59 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.42 CEBPBCCAAT/enhancer binding protein (C/EBP), beta (212501_at), score: 0.33 CFBcomplement factor B (202357_s_at), score: 0.38 CG012hypothetical gene CG012 (215318_at), score: 0.57 CHI3L1chitinase 3-like 1 (cartilage glycoprotein-39) (209395_at), score: 0.37 CHST2carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 (203921_at), score: 0.5 CHST7carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 7 (206756_at), score: -0.59 CKBcreatine kinase, brain (200884_at), score: -0.56 CLDN18claudin 18 (214135_at), score: 0.4 CLEC11AC-type lectin domain family 11, member A (211709_s_at), score: -0.64 CLEC2BC-type lectin domain family 2, member B (209732_at), score: 0.4 CLGNcalmegin (205830_at), score: 0.55 CLIC2chloride intracellular channel 2 (213415_at), score: 0.44 CLUclusterin (208791_at), score: 0.6 CMAHcytidine monophosphate-N-acetylneuraminic acid hydroxylase (CMP-N-acetylneuraminate monooxygenase) pseudogene (205518_s_at), score: 0.34 COASYCoenzyme A synthase (201913_s_at), score: -0.56 COL15A1collagen, type XV, alpha 1 (203477_at), score: 0.37 COL16A1collagen, type XVI, alpha 1 (204345_at), score: 0.34 COX7A2cytochrome c oxidase subunit VIIa polypeptide 2 (liver) (217249_x_at), score: -0.59 CPMcarboxypeptidase M (206100_at), score: 0.37 CPZcarboxypeptidase Z (211062_s_at), score: 0.47 CREG1cellular repressor of E1A-stimulated genes 1 (201200_at), score: 0.37 CScitrate synthase (208660_at), score: -0.57 CST6cystatin E/M (206595_at), score: -0.58 CSTAcystatin A (stefin A) (204971_at), score: 0.4 CTTNBP2NLCTTNBP2 N-terminal like (214731_at), score: 0.58 CXCL2chemokine (C-X-C motif) ligand 2 (209774_x_at), score: 0.43 CXCL3chemokine (C-X-C motif) ligand 3 (207850_at), score: 0.34 CXCL6chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) (206336_at), score: 0.45 CXCR7chemokine (C-X-C motif) receptor 7 (212977_at), score: 0.59 CXorf21chromosome X open reading frame 21 (220252_x_at), score: 0.35 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.54 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: 0.52 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (207386_at), score: 0.32 DAPK1death-associated protein kinase 1 (203139_at), score: 0.48 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (219290_x_at), score: 0.39 DBC1deleted in bladder cancer 1 (205818_at), score: 0.92 DDX21DEAD (Asp-Glu-Ala-Asp) box polypeptide 21 (208152_s_at), score: 0.33 DHODHdihydroorotate dehydrogenase (213631_x_at), score: 0.36 DHRS1dehydrogenase/reductase (SDR family) member 1 (213279_at), score: -0.6 DIP2ADIP2 disco-interacting protein 2 homolog A (Drosophila) (215529_x_at), score: 0.42 DKFZP586I1420hypothetical protein DKFZp586I1420 (213546_at), score: 0.47 DKK2dickkopf homolog 2 (Xenopus laevis) (219908_at), score: 0.8 DLGAP4discs, large (Drosophila) homolog-associated protein 4 (202570_s_at), score: -0.61 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.38 DNM3dynamin 3 (209839_at), score: 0.33 DPP3dipeptidyl-peptidase 3 (218567_x_at), score: -0.6 DPP4dipeptidyl-peptidase 4 (211478_s_at), score: 0.51 DPTdermatopontin (213068_at), score: 0.83 DYMdymeclin (220774_at), score: 0.32 DYNC2H1dynein, cytoplasmic 2, heavy chain 1 (219469_at), score: 0.35 EDAectodysplasin A (211130_x_at), score: 0.51 EGFL6EGF-like-domain, multiple 6 (219454_at), score: 0.83 EIF4A2eukaryotic translation initiation factor 4A, isoform 2 (200912_s_at), score: -0.57 ELNelastin (212670_at), score: 0.38 ENDOD1endonuclease domain containing 1 (212573_at), score: -0.69 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.32 ENPP4ectonucleotide pyrophosphatase/phosphodiesterase 4 (putative function) (204160_s_at), score: 0.54 EPHB6EPH receptor B6 (204718_at), score: 0.4 ERAP2endoplasmic reticulum aminopeptidase 2 (219759_at), score: 0.35 FAM106Afamily with sequence similarity 106, member A (220575_at), score: 0.31 FAM149B1family with sequence similarity 149, member B1 (213896_x_at), score: -0.58 FAM174Bfamily with sequence similarity 174, member B (51158_at), score: 0.39 FAPfibroblast activation protein, alpha (209955_s_at), score: 0.51 FAT4FAT tumor suppressor homolog 4 (Drosophila) (219427_at), score: 0.56 FBLN1fibulin 1 (202995_s_at), score: 0.56 FBLN2fibulin 2 (203886_s_at), score: 0.47 FBXL5F-box and leucine-rich repeat protein 5 (209004_s_at), score: -0.71 FCGRTFc fragment of IgG, receptor, transporter, alpha (218831_s_at), score: 0.42 FGF13fibroblast growth factor 13 (205110_s_at), score: 0.65 FGF5fibroblast growth factor 5 (210310_s_at), score: -0.57 FGF9fibroblast growth factor 9 (glia-activating factor) (206404_at), score: 0.67 FLGfilaggrin (215704_at), score: 0.34 FLJ42627hypothetical LOC645644 (220352_x_at), score: 0.37 FLRT3fibronectin leucine rich transmembrane protein 3 (219250_s_at), score: 0.57 FMODfibromodulin (202709_at), score: 0.47 FRAT1frequently rearranged in advanced T-cell lymphomas (219889_at), score: 0.39 FTHP1ferritin, heavy polypeptide pseudogene 1 (211628_x_at), score: 0.39 FUBP1far upstream element (FUSE) binding protein 1 (212847_at), score: 0.33 GABBR1gamma-aminobutyric acid (GABA) B receptor, 1 (203146_s_at), score: 0.52 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (204537_s_at), score: 0.7 GALNT10UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 10 (GalNAc-T10) (207357_s_at), score: -0.62 GALNT6UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 6 (GalNAc-T6) (219956_at), score: -0.58 GAS7growth arrest-specific 7 (202191_s_at), score: 0.34 GCH1GTP cyclohydrolase 1 (204224_s_at), score: 0.39 GGHgamma-glutamyl hydrolase (conjugase, folylpolygammaglutamyl hydrolase) (203560_at), score: -0.65 GJA1gap junction protein, alpha 1, 43kDa (201667_at), score: 0.36 GM2AGM2 ganglioside activator (212737_at), score: -0.62 GOLSYNGolgi-localized protein (218692_at), score: 0.75 GPNMBglycoprotein (transmembrane) nmb (201141_at), score: 0.35 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: 0.34 GRIA1glutamate receptor, ionotropic, AMPA 1 (209793_at), score: 0.75 GRIA3glutamate receptor, ionotrophic, AMPA 3 (206730_at), score: 0.36 GRM6glutamate receptor, metabotropic 6 (208035_at), score: 0.36 GSTA1glutathione S-transferase alpha 1 (215766_at), score: 0.36 GYPCglycophorin C (Gerbich blood group) (202947_s_at), score: 0.38 HCG8HLA complex group 8 (215985_at), score: 0.34 HMOX1heme oxygenase (decycling) 1 (203665_at), score: 0.5 HNMThistamine N-methyltransferase (204112_s_at), score: 0.4 HOXA5homeobox A5 (213844_at), score: 0.41 HOXA7homeobox A7 (206848_at), score: 0.5 HYMAIhydatidiform mole associated and imprinted (non-protein coding) (215513_at), score: 0.39 IBD12Inflammatory bowel disease 12 (215373_x_at), score: 0.33 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.38 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.35 IGF1insulin-like growth factor 1 (somatomedin C) (209541_at), score: 0.55 IGFBP7insulin-like growth factor binding protein 7 (201162_at), score: -0.67 IL15RAinterleukin 15 receptor, alpha (207375_s_at), score: 0.4 IL18R1interleukin 18 receptor 1 (206618_at), score: 0.5 IL1R1interleukin 1 receptor, type I (202948_at), score: 0.33 IL1RAPinterleukin 1 receptor accessory protein (205227_at), score: 0.33 IL1RL1interleukin 1 receptor-like 1 (207526_s_at), score: 0.86 IL7interleukin 7 (206693_at), score: 0.68 IMAASLC7A5 pseudogene (208118_x_at), score: 0.41 INE1inactivation escape 1 (non-protein coding) (207252_at), score: 0.32 IRAK3interleukin-1 receptor-associated kinase 3 (213817_at), score: 0.48 ITGA3integrin, alpha 3 (antigen CD49C, alpha 3 subunit of VLA-3 receptor) (201474_s_at), score: -0.62 ITGB8integrin, beta 8 (211488_s_at), score: 0.59 ITIH5inter-alpha (globulin) inhibitor H5 (219064_at), score: 1 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: 0.43 JHDM1Djumonji C domain containing histone demethylase 1 homolog D (S. cerevisiae) (221778_at), score: 0.44 JMJD3jumonji domain containing 3, histone lysine demethylase (213146_at), score: 0.49 JMJD7jumonji domain containing 7 (60528_at), score: 0.45 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (206765_at), score: 0.35 KCNK1potassium channel, subfamily K, member 1 (204679_at), score: 0.33 KIAA0644KIAA0644 gene product (205150_s_at), score: 0.35 KIAA0894KIAA0894 protein (207436_x_at), score: 0.38 KIAA1644KIAA1644 (52837_at), score: 0.59 KLF5Kruppel-like factor 5 (intestinal) (209212_s_at), score: -0.57 KRT19keratin 19 (201650_at), score: -0.6 LANCL1LanC lantibiotic synthetase component C-like 1 (bacterial) (202020_s_at), score: -0.56 LAPTM4Blysosomal protein transmembrane 4 beta (208767_s_at), score: -0.58 LAX1lymphocyte transmembrane adaptor 1 (207734_at), score: 0.4 LGMNlegumain (201212_at), score: -0.57 LHPPphospholysine phosphohistidine inorganic pyrophosphate phosphatase (218523_at), score: -0.6 LOC100132134similar to LOC653391 protein (213089_at), score: 0.4 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.4 LOC100133748similar to GTF2IRD2 protein (215569_at), score: 0.32 LOC100188945cell division cycle associated 4 pseudogene (215109_at), score: 0.35 LOC149478hypothetical protein LOC149478 (215462_at), score: 0.5 LOC150759hypothetical protein LOC150759 (213703_at), score: 0.45 LOC23117PI-3-kinase-related kinase SMG-1 isoform 1 homolog (211996_s_at), score: 0.48 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.34 LOC399491LOC399491 protein (214035_x_at), score: 0.41 LOC643293FLJ44451 pseudogene (215057_at), score: 0.41 LOC647070hypothetical LOC647070 (215467_x_at), score: 0.32 LOC728844hypothetical LOC728844 (222040_at), score: 0.34 LOC729806similar to hCG1725380 (217544_at), score: 0.43 LOC791120hypothetical LOC791120 (213367_at), score: 0.54 LOH3CR2Aloss of heterozygosity, 3, chromosomal region 2, gene A (220244_at), score: 0.49 LPCAT4lysophosphatidylcholine acyltransferase 4 (40472_at), score: -0.69 LRP1low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) (200784_s_at), score: 0.49 LUZP1leucine zipper protein 1 (221832_s_at), score: -0.59 MAFBv-maf musculoaponeurotic fibrosarcoma oncogene homolog B (avian) (218559_s_at), score: 0.52 MAN1A1mannosidase, alpha, class 1A, member 1 (221760_at), score: 0.45 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.47 MAN2C1mannosidase, alpha, class 2C, member 1 (203668_at), score: 0.42 MAP2K6mitogen-activated protein kinase kinase 6 (205698_s_at), score: 0.41 MAP3K4mitogen-activated protein kinase kinase kinase 4 (216199_s_at), score: 0.35 MATN2matrilin 2 (202350_s_at), score: -0.59 MCCmutated in colorectal cancers (206132_at), score: 0.39 MEIS3P1Meis homeobox 3 pseudogene 1 (214077_x_at), score: 0.36 MFAP4microfibrillar-associated protein 4 (212713_at), score: 0.46 MKNK1MAP kinase interacting serine/threonine kinase 1 (209467_s_at), score: 0.34 MKRN1makorin ring finger protein 1 (208082_x_at), score: 0.34 MLLmyeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila) (212080_at), score: 0.31 MLLT11myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 11 (211071_s_at), score: -0.69 MMEmembrane metallo-endopeptidase (203434_s_at), score: 0.33 MRPL49mitochondrial ribosomal protein L49 (201717_at), score: -0.6 MSCmusculin (activated B-cell factor-1) (209928_s_at), score: 0.52 MUC3Amucin 3A, cell surface associated (217117_x_at), score: 0.38 MYO1Amyosin IA (211916_s_at), score: 0.35 MYO1Dmyosin ID (212338_at), score: 0.5 NACAnascent polypeptide-associated complex alpha subunit (222018_at), score: 0.44 NARG1LNMDA receptor regulated 1-like (219378_at), score: 0.36 NBEAneurobeachin (221207_s_at), score: 0.49 NCRNA00084non-protein coding RNA 84 (214657_s_at), score: 0.43 NDRG3NDRG family member 3 (217286_s_at), score: -0.57 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: -0.58 NDUFA7NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa (202785_at), score: -0.62 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (210162_s_at), score: 0.34 NOTCH3Notch homolog 3 (Drosophila) (203238_s_at), score: -0.64 NOVA1neuro-oncological ventral antigen 1 (205794_s_at), score: 0.39 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.35 NPTX2neuronal pentraxin II (213479_at), score: 0.35 NR3C1nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) (201866_s_at), score: 0.39 NRCAMneuronal cell adhesion molecule (204105_s_at), score: 0.73 OBFC1oligonucleotide/oligosaccharide-binding fold containing 1 (219100_at), score: -0.61 OCEL1occludin/ELL domain containing 1 (205441_at), score: -0.67 OR2A9Polfactory receptor, family 2, subfamily A, member 9 pseudogene (222290_at), score: 0.32 ORAI2ORAI calcium release-activated calcium modulator 2 (218812_s_at), score: 0.33 PAAF1proteasomal ATPase-associated factor 1 (218957_s_at), score: -0.6 PALLDpalladin, cytoskeletal associated protein (200906_s_at), score: -0.58 PALMparalemmin (203859_s_at), score: 0.39 PAPPApregnancy-associated plasma protein A, pappalysin 1 (201981_at), score: 0.41 PAPPA2pappalysin 2 (213332_at), score: 0.45 PCDHGA8protocadherin gamma subfamily A, 8 (210368_at), score: 0.38 PCSK5proprotein convertase subtilisin/kexin type 5 (213652_at), score: 0.48 PDGFDplatelet derived growth factor D (219304_s_at), score: 0.67 PDGFRLplatelet-derived growth factor receptor-like (205226_at), score: 0.49 PDPNpodoplanin (221898_at), score: 0.46 PERPPERP, TP53 apoptosis effector (217744_s_at), score: -0.57 PFN2profilin 2 (204992_s_at), score: -0.7 PHC3polyhomeotic homolog 3 (Drosophila) (215521_at), score: 0.49 PHF20L1PHD finger protein 20-like 1 (222133_s_at), score: 0.39 PID1phosphotyrosine interaction domain containing 1 (219093_at), score: 0.34 PIGFphosphatidylinositol glycan anchor biosynthesis, class F (205078_at), score: -0.59 PLEKHB2pleckstrin homology domain containing, family B (evectins) member 2 (201411_s_at), score: -0.59 PLGLB1plasminogen-like B1 (214415_at), score: 0.51 PLGLB2plasminogen-like B2 (205871_at), score: 0.63 PLSCR4phospholipid scramblase 4 (218901_at), score: 0.38 PLXNB1plexin B1 (215807_s_at), score: 0.4 POGZpogo transposable element with ZNF domain (215281_x_at), score: 0.33 POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (220113_x_at), score: 0.38 POLR2J4polymerase (RNA) II (DNA directed) polypeptide J4, pseudogene (60815_at), score: 0.59 PPLperiplakin (203407_at), score: 0.75 PROS1protein S (alpha) (207808_s_at), score: 0.36 PRR5proline rich 5 (renal) (47069_at), score: 0.34 PTBP2polypyrimidine tract binding protein 2 (218683_at), score: 0.31 PTPLAprotein tyrosine phosphatase-like (proline instead of catalytic arginine), member A (219654_at), score: -0.57 PTPREprotein tyrosine phosphatase, receptor type, E (221840_at), score: 0.43 PTTG1pituitary tumor-transforming 1 (203554_x_at), score: -0.61 RALGPS2Ral GEF with PH domain and SH3 binding motif 2 (220338_at), score: 0.59 RBM39RNA binding motif protein 39 (207941_s_at), score: 0.35 RCAN2regulator of calcineurin 2 (203498_at), score: 0.44 RECQL5RecQ protein-like 5 (34063_at), score: 0.36 RGPD5RANBP2-like and GRIP domain containing 5 (210676_x_at), score: 0.41 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: 0.32 RPAP2RNA polymerase II associated protein 2 (219504_s_at), score: 0.4 RPL23AP32ribosomal protein L23a pseudogene 32 (207283_at), score: 0.37 RPL26L1ribosomal protein L26-like 1 (218830_at), score: -0.6 RPS20ribosomal protein S20 (216247_at), score: 0.32 RUFY2RUN and FYVE domain containing 2 (219957_at), score: 0.31 SEMA5Asema domain, seven thrombospondin repeats (type 1 and type 1-like), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 5A (205405_at), score: 0.4 SENP6SUMO1/sentrin specific peptidase 6 (202318_s_at), score: 0.34 SEPP1selenoprotein P, plasma, 1 (201427_s_at), score: 0.74 SEPW1selenoprotein W, 1 (201194_at), score: -0.62 SERPINE2serpin peptidase inhibitor, clade E (nexin, plasminogen activator inhibitor type 1), member 2 (212190_at), score: 0.32 SERPINF1serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 (202283_at), score: 0.4 SETBP1SET binding protein 1 (205933_at), score: 0.32 SETMARSET domain and mariner transposase fusion gene (206554_x_at), score: 0.36 SFRS11splicing factor, arginine/serine-rich 11 (213742_at), score: 0.36 SH2B2SH2B adaptor protein 2 (205367_at), score: 0.36 SH2D3ASH2 domain containing 3A (222169_x_at), score: 0.33 SKP1S-phase kinase-associated protein 1 (200719_at), score: 0.4 SLC12A7solute carrier family 12 (potassium/chloride transporters), member 7 (218066_at), score: 0.33 SLC17A5solute carrier family 17 (anion/sugar transporter), member 5 (221041_s_at), score: -0.64 SLC1A3solute carrier family 1 (glial high affinity glutamate transporter), member 3 (202800_at), score: 0.39 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (218749_s_at), score: 0.32 SLC25A37solute carrier family 25, member 37 (218136_s_at), score: 0.37 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: 0.38 SLC39A8solute carrier family 39 (zinc transporter), member 8 (209267_s_at), score: 0.61 SLC7A7solute carrier family 7 (cationic amino acid transporter, y+ system), member 7 (204588_s_at), score: 0.35 SLIT2slit homolog 2 (Drosophila) (209897_s_at), score: 0.33 SLIT3slit homolog 3 (Drosophila) (203813_s_at), score: 0.47 SMA4glucuronidase, beta pseudogene (215599_at), score: 0.42 SMOXspermine oxidase (210357_s_at), score: 0.36 SMPD1sphingomyelin phosphodiesterase 1, acid lysosomal (209420_s_at), score: -0.58 SMPDL3Asphingomyelin phosphodiesterase, acid-like 3A (213624_at), score: 0.32 SNED1sushi, nidogen and EGF-like domains 1 (213488_at), score: 0.42 SOD2superoxide dismutase 2, mitochondrial (221477_s_at), score: 0.35 SOX4SRY (sex determining region Y)-box 4 (201417_at), score: 0.39 SPG21spastic paraplegia 21 (autosomal recessive, Mast syndrome) (215383_x_at), score: 0.36 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.77 SPRED2sprouty-related, EVH1 domain containing 2 (212458_at), score: 0.37 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.36 SQSTM1sequestosome 1 (201471_s_at), score: 0.37 STMN1stathmin 1/oncoprotein 18 (200783_s_at), score: -0.58 SUPT3Hsuppressor of Ty 3 homolog (S. cerevisiae) (206506_s_at), score: -0.59 SUPT4H1suppressor of Ty 4 homolog 1 (S. cerevisiae) (201484_at), score: -0.59 SVEP1sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 (213247_at), score: 0.56 SVILsupervillin (202565_s_at), score: 0.53 SWAP70SWAP-70 protein (209307_at), score: 0.41 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: 0.32 TAP1transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) (202307_s_at), score: -0.6 TBC1D1TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 (212350_at), score: -0.61 TBC1D3TBC1 domain family, member 3 (209403_at), score: 0.37 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.32 TCTN3tectonic family member 3 (212121_at), score: -0.66 TDRD7tudor domain containing 7 (213361_at), score: -0.58 THBDthrombomodulin (203887_s_at), score: 0.33 TIGD1Ltigger transposable element derived 1-like (216459_x_at), score: 0.34 TJP2tight junction protein 2 (zona occludens 2) (202085_at), score: -0.61 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.57 TMEM176Btransmembrane protein 176B (220532_s_at), score: 0.7 TMSB10thymosin beta 10 (217733_s_at), score: -0.56 TNCtenascin C (201645_at), score: 0.39 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.52 TNFRSF11Btumor necrosis factor receptor superfamily, member 11b (204933_s_at), score: 0.36 TNFSF10tumor necrosis factor (ligand) superfamily, member 10 (214329_x_at), score: 0.41 TNS3tensin 3 (217853_at), score: 0.36 TP53I11tumor protein p53 inducible protein 11 (214667_s_at), score: 0.45 TPMTthiopurine S-methyltransferase (203672_x_at), score: -0.61 TPST1tyrosylprotein sulfotransferase 1 (204140_at), score: 0.36 TRA2Atransformer 2 alpha homolog (Drosophila) (213593_s_at), score: 0.32 TREX1three prime repair exonuclease 1 (34689_at), score: -0.56 TRIM66tripartite motif-containing 66 (213748_at), score: 0.43 TRPC6transient receptor potential cation channel, subfamily C, member 6 (217287_s_at), score: 0.33 TRPS1trichorhinophalangeal syndrome I (218502_s_at), score: 0.35 TYMPthymidine phosphorylase (217497_at), score: 0.38 UBE2D2ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) (201345_s_at), score: -0.69 UBQLN4ubiquilin 4 (222252_x_at), score: 0.37 ULBP1UL16 binding protein 1 (221323_at), score: 0.4 USP12ubiquitin specific peptidase 12 (213327_s_at), score: 0.34 USP6ubiquitin specific peptidase 6 (Tre-2 oncogene) (206405_x_at), score: 0.35 VDRvitamin D (1,25- dihydroxyvitamin D3) receptor (204255_s_at), score: 0.59 VEGFAvascular endothelial growth factor A (211527_x_at), score: 0.33 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (214785_at), score: 0.36 WISP1WNT1 inducible signaling pathway protein 1 (206796_at), score: 0.69 WISP2WNT1 inducible signaling pathway protein 2 (205792_at), score: 0.72 WTAPWilms tumor 1 associated protein (210285_x_at), score: 0.4 XISTX (inactive)-specific transcript (non-protein coding) (214218_s_at), score: 0.37 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.34 ZC3H7Bzinc finger CCCH-type containing 7B (206169_x_at), score: 0.44 ZMIZ1zinc finger, MIZ-type containing 1 (212124_at), score: 0.46 ZNF136zinc finger protein 136 (206240_s_at), score: 0.4 ZNF160zinc finger protein 160 (214715_x_at), score: 0.33 ZNF197zinc finger protein 197 (205855_at), score: 0.34 ZNF219zinc finger protein 219 (219314_s_at), score: 0.45 ZNF224zinc finger protein 224 (216983_s_at), score: 0.44 ZNF37Bzinc finger protein 37B (215358_x_at), score: 0.37 ZNF551zinc finger protein 551 (211721_s_at), score: 0.33 ZNF614zinc finger protein 614 (220721_at), score: 0.32 ZNF652zinc finger protein 652 (205594_at), score: 0.35 ZNF767zinc finger family member 767 (219627_at), score: 0.35
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| F055_WBS.CEL | 14 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F348_WBS.CEL | 16 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| 4319_WBS.CEL | 5 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| D890_WBS.CEL | 13 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| F223_WBS.CEL | 15 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| ctrl c 08-03.CEL | 3 | 1 | DS-CC | hgu133a | none | DS-CC 3 |
| 2433_CNTL.CEL | 4 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| 8495_CNTL.CEL | 10 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl b 08-03.CEL | 2 | 1 | DS-CC | hgu133a | none | DS-CC 2 |
| 9118_CNTL.CEL | 11 | 8 | WBS | hgu133plus2 | none | WBS 1 |
| ctrl a 08-03.CEL | 1 | 1 | DS-CC | hgu133a | none | DS-CC 1 |