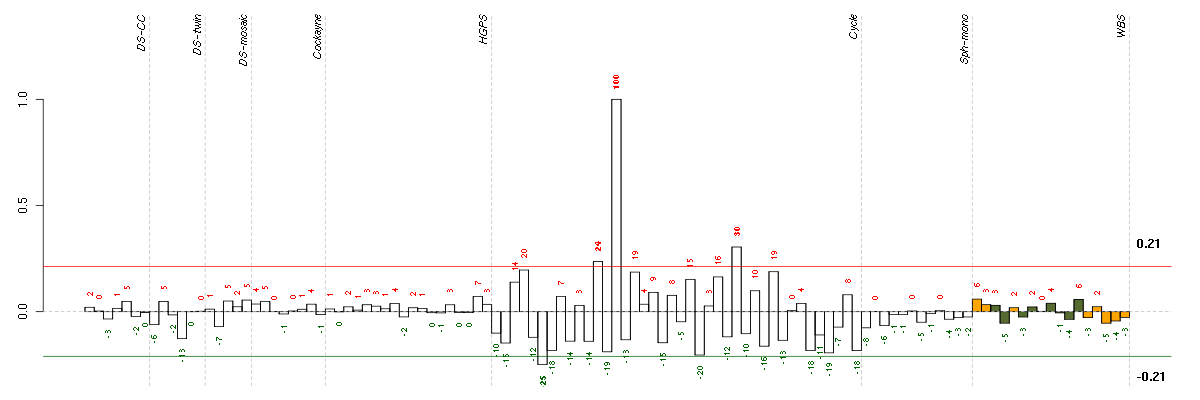

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
This term is the most general term possible
ABCC4ATP-binding cassette, sub-family C (CFTR/MRP), member 4 (203196_at), score: -0.51 ABHD11abhydrolase domain containing 11 (221927_s_at), score: 0.38 ABI1abl-interactor 1 (209028_s_at), score: -0.58 ABT1activator of basal transcription 1 (218405_at), score: -0.53 ABTB2ankyrin repeat and BTB (POZ) domain containing 2 (213497_at), score: -0.65 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: -0.51 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.45 ACSL6acyl-CoA synthetase long-chain family member 6 (211207_s_at), score: 0.66 ACTN3actinin, alpha 3 (206891_at), score: -0.72 ADAMDEC1ADAM-like, decysin 1 (206134_at), score: 0.78 ADAMTS7ADAM metallopeptidase with thrombospondin type 1 motif, 7 (220705_s_at), score: -0.47 ADAMTS9ADAM metallopeptidase with thrombospondin type 1 motif, 9 (220287_at), score: 0.63 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.49 AGAP2ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 (215080_s_at), score: 0.57 AHCYL2S-adenosylhomocysteine hydrolase-like 2 (215672_s_at), score: -0.58 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (207102_at), score: 0.42 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (221589_s_at), score: -0.47 AMBRA1autophagy/beclin-1 regulator 1 (52731_at), score: -0.46 AMTaminomethyltransferase (204294_at), score: -0.49 ANGEL1angel homolog 1 (Drosophila) (36865_at), score: -0.58 ANKS1Aankyrin repeat and sterile alpha motif domain containing 1A (212747_at), score: -0.58 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (204650_s_at), score: -0.44 APOC1apolipoprotein C-I (213553_x_at), score: -0.64 APOL6apolipoprotein L, 6 (219716_at), score: -0.44 APPL1adaptor protein, phosphotyrosine interaction, PH domain and leucine zipper containing 1 (218158_s_at), score: -0.61 ARHGAP5Rho GTPase activating protein 5 (217936_at), score: -0.47 ARL15ADP-ribosylation factor-like 15 (219842_at), score: -0.5 ASTN2astrotactin 2 (209693_at), score: -0.53 ATAD5ATPase family, AAA domain containing 5 (220223_at), score: 0.36 ATF1activating transcription factor 1 (222103_at), score: -0.45 ATHL1ATH1, acid trehalase-like 1 (yeast) (219359_at), score: 0.57 ATMataxia telangiectasia mutated (210858_x_at), score: -0.57 ATP11AATPase, class VI, type 11A (215842_s_at), score: -0.46 ATP1B4ATPase, (Na+)/K+ transporting, beta 4 polypeptide (220556_at), score: 0.43 ATP8A1ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 (213106_at), score: 0.36 ATP8B2ATPase, class I, type 8B, member 2 (216873_s_at), score: -0.51 ATPAF2ATP synthase mitochondrial F1 complex assembly factor 2 (213057_at), score: -0.46 ATXN1ataxin 1 (203231_s_at), score: -0.45 ATXN7ataxin 7 (204516_at), score: -0.62 AVILadvillin (205539_at), score: -0.66 BAIAP3BAI1-associated protein 3 (216356_x_at), score: -0.43 BBS10Bardet-Biedl syndrome 10 (219487_at), score: -0.6 BCL2L1BCL2-like 1 (215037_s_at), score: 0.42 BEANbrain expressed, associated with Nedd4 (214068_at), score: 0.36 BICC1bicaudal C homolog 1 (Drosophila) (220580_at), score: 0.41 BNC1basonuclin 1 (206581_at), score: -0.44 BRAFv-raf murine sarcoma viral oncogene homolog B1 (206044_s_at), score: 0.48 BTBD7BTB (POZ) domain containing 7 (220297_at), score: -0.51 C10orf118chromosome 10 open reading frame 118 (219844_at), score: -0.6 C10orf72chromosome 10 open reading frame 72 (213381_at), score: -0.45 C14orf135chromosome 14 open reading frame 135 (219972_s_at), score: -0.52 C14orf138chromosome 14 open reading frame 138 (218940_at), score: -0.5 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.44 C16orf7chromosome 16 open reading frame 7 (205781_at), score: -0.44 C1orf115chromosome 1 open reading frame 115 (218546_at), score: 0.38 C1orf38chromosome 1 open reading frame 38 (207571_x_at), score: -0.45 C1orf66chromosome 1 open reading frame 66 (218914_at), score: -0.49 C1QTNF1C1q and tumor necrosis factor related protein 1 (220975_s_at), score: -0.59 C20orf111chromosome 20 open reading frame 111 (209020_at), score: -0.53 C20orf12chromosome 20 open reading frame 12 (219951_s_at), score: -0.52 C21orf2chromosome 21 open reading frame 2 (203996_s_at), score: -0.53 C3AR1complement component 3a receptor 1 (209906_at), score: -0.6 C3orf51chromosome 3 open reading frame 51 (208247_at), score: 0.51 C4orf16chromosome 4 open reading frame 16 (219023_at), score: -0.52 C4orf29chromosome 4 open reading frame 29 (219980_at), score: 0.65 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.64 C6orf47chromosome 6 open reading frame 47 (204968_at), score: -0.44 C6orf64chromosome 6 open reading frame 64 (218784_s_at), score: -0.48 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.52 C8orf41chromosome 8 open reading frame 41 (219124_at), score: 0.36 C8orf44chromosome 8 open reading frame 44 (220216_at), score: -0.52 C9orf7chromosome 9 open reading frame 7 (61874_at), score: -0.52 CAPN5calpain 5 (205166_at), score: 0.38 CC2D1Acoiled-coil and C2 domain containing 1A (58994_at), score: -0.44 CCDC101coiled-coil domain containing 101 (48117_at), score: -0.5 CCDC121coiled-coil domain containing 121 (220321_s_at), score: 0.48 CCRKcell cycle related kinase (205271_s_at), score: -0.43 CD200CD200 molecule (209583_s_at), score: 0.57 CD28CD28 molecule (206545_at), score: 0.65 CD40CD40 molecule, TNF receptor superfamily member 5 (215346_at), score: -0.53 CD53CD53 molecule (203416_at), score: -0.49 CDC14BCDC14 cell division cycle 14 homolog B (S. cerevisiae) (208022_s_at), score: -0.46 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.35 CDH19cadherin 19, type 2 (206898_at), score: 1 CDK10cyclin-dependent kinase 10 (203468_at), score: -0.49 CDRT1CMT1A duplicated region transcript 1 (215999_at), score: -0.43 CHKAcholine kinase alpha (204233_s_at), score: -0.45 CHN2chimerin (chimaerin) 2 (207486_x_at), score: 0.56 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.46 CHRNA5cholinergic receptor, nicotinic, alpha 5 (206533_at), score: 0.53 CIRCBF1 interacting corepressor (209571_at), score: -0.45 CLCN2chloride channel 2 (213499_at), score: -0.46 CLEC1AC-type lectin domain family 1, member A (219761_at), score: 0.41 CLTCL1clathrin, heavy chain-like 1 (205944_s_at), score: -0.61 CNKSR1connector enhancer of kinase suppressor of Ras 1 (204740_at), score: -0.51 CNNM1cyclin M1 (220166_at), score: -0.53 CNR2cannabinoid receptor 2 (macrophage) (206586_at), score: -0.44 CNTLNcentlein, centrosomal protein (220095_at), score: 0.44 COL10A1collagen, type X, alpha 1 (217428_s_at), score: 0.49 COL16A1collagen, type XVI, alpha 1 (204345_at), score: -0.43 COPS7BCOP9 constitutive photomorphogenic homolog subunit 7B (Arabidopsis) (219997_s_at), score: -0.43 COQ7coenzyme Q7 homolog, ubiquinone (yeast) (209745_at), score: 0.5 COX6A2cytochrome c oxidase subunit VIa polypeptide 2 (206353_at), score: -0.59 CPA2carboxypeptidase A2 (pancreatic) (206212_at), score: -0.43 CPA4carboxypeptidase A4 (205832_at), score: -0.46 CPNE3copine III (202118_s_at), score: 0.36 CPT2carnitine palmitoyltransferase 2 (204264_at), score: -0.56 CRHR1corticotropin releasing hormone receptor 1 (208593_x_at), score: -0.56 CRY1cryptochrome 1 (photolyase-like) (209674_at), score: -0.47 CSPG5chondroitin sulfate proteoglycan 5 (neuroglycan C) (39966_at), score: -0.52 CSTF2Tcleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa, tau variant (212905_at), score: 0.45 CTAGE4CTAGE family, member 4 (215549_x_at), score: -0.5 CTSScathepsin S (202901_x_at), score: 0.93 CXCL11chemokine (C-X-C motif) ligand 11 (210163_at), score: 0.62 CYCSP52cytochrome c, somatic pseudogene 52 (217206_at), score: 0.76 CYP24A1cytochrome P450, family 24, subfamily A, polypeptide 1 (206504_at), score: 0.38 CYP26B1cytochrome P450, family 26, subfamily B, polypeptide 1 (219825_at), score: 0.53 CYP2C9cytochrome P450, family 2, subfamily C, polypeptide 9 (214421_x_at), score: -0.43 CYP3A43cytochrome P450, family 3, subfamily A, polypeptide 43 (211440_x_at), score: -0.43 CYSLTR1cysteinyl leukotriene receptor 1 (216288_at), score: 0.53 DCUN1D2DCN1, defective in cullin neddylation 1, domain containing 2 (S. cerevisiae) (219116_s_at), score: -0.54 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (218579_s_at), score: 0.43 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.56 DMXL2Dmx-like 2 (212820_at), score: -0.48 DNAL4dynein, axonemal, light chain 4 (204008_at), score: -0.46 DRD5dopamine receptor D5 (208486_at), score: -0.43 DSC2desmocollin 2 (204751_x_at), score: 0.4 DSCR6Down syndrome critical region gene 6 (207267_s_at), score: 0.81 DSG2desmoglein 2 (217901_at), score: 0.4 DSPPdentin sialophosphoprotein (221681_s_at), score: -0.54 DSTYKdual serine/threonine and tyrosine protein kinase (214663_at), score: -0.51 DTNAdystrobrevin, alpha (205741_s_at), score: 0.47 DTNBdystrobrevin, beta (215295_at), score: 0.83 DTX2deltex homolog 2 (Drosophila) (215732_s_at), score: 0.55 DUSP8dual specificity phosphatase 8 (206374_at), score: -0.48 DYMdymeclin (220774_at), score: -0.47 DYNC2H1dynein, cytoplasmic 2, heavy chain 1 (219469_at), score: -0.43 EBPemopamil binding protein (sterol isomerase) (202735_at), score: -0.43 EDA2Rectodysplasin A2 receptor (221399_at), score: 0.55 EFNB1ephrin-B1 (202711_at), score: -0.52 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.38 EIF2AK2eukaryotic translation initiation factor 2-alpha kinase 2 (204211_x_at), score: -0.53 EIF2C4eukaryotic translation initiation factor 2C, 4 (219190_s_at), score: -0.51 EIF4ENIF1eukaryotic translation initiation factor 4E nuclear import factor 1 (218626_at), score: -0.46 ELA3Aelastase 3A, pancreatic (211738_x_at), score: -0.53 ELAC1elaC homolog 1 (E. coli) (219325_s_at), score: -0.43 ELLelongation factor RNA polymerase II (204096_s_at), score: -0.59 EML2echinoderm microtubule associated protein like 2 (204398_s_at), score: -0.62 ENPEPglutamyl aminopeptidase (aminopeptidase A) (204844_at), score: 0.37 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.5 EPHA3EPH receptor A3 (206070_s_at), score: 0.36 EPHB6EPH receptor B6 (204718_at), score: 0.37 EPM2Aepilepsy, progressive myoclonus type 2A, Lafora disease (laforin) (205231_s_at), score: -0.55 EPOerythropoietin (217254_s_at), score: -0.52 ERBB3v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) (202454_s_at), score: 0.41 ERBB4v-erb-a erythroblastic leukemia viral oncogene homolog 4 (avian) (214053_at), score: 0.7 ERMAPerythroblast membrane-associated protein (Scianna blood group) (219905_at), score: 0.38 ERN1endoplasmic reticulum to nucleus signaling 1 (207061_at), score: 0.41 EXD3exonuclease 3'-5' domain containing 3 (220838_at), score: -0.57 EXOSC5exosome component 5 (218481_at), score: -0.48 F10coagulation factor X (205620_at), score: -0.46 F7coagulation factor VII (serum prothrombin conversion accelerator) (207300_s_at), score: -0.44 FA2Hfatty acid 2-hydroxylase (219429_at), score: 0.39 FAM169Afamily with sequence similarity 169, member A (213954_at), score: 0.48 FAM173Afamily with sequence similarity 173, member A (219709_x_at), score: -0.49 FBRSfibrosin (218255_s_at), score: -0.48 FBXO17F-box protein 17 (220233_at), score: 0.48 FBXO2F-box protein 2 (219305_x_at), score: -0.59 FBXO22F-box protein 22 (219638_at), score: 0.67 FBXO46F-box protein 46 (205310_at), score: -0.49 FCER1AFc fragment of IgE, high affinity I, receptor for; alpha polypeptide (211734_s_at), score: 0.43 FCGR2AFc fragment of IgG, low affinity IIa, receptor (CD32) (203561_at), score: 0.49 FER1L4fer-1-like 4 (C. elegans) (222245_s_at), score: -0.51 FGL1fibrinogen-like 1 (205305_at), score: -0.43 FKRPfukutin related protein (219853_at), score: 0.55 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.48 FMO3flavin containing monooxygenase 3 (40665_at), score: 0.41 FMR1fragile X mental retardation 1 (215245_x_at), score: -0.52 FNBP1Lformin binding protein 1-like (215017_s_at), score: -0.53 FOXE1forkhead box E1 (thyroid transcription factor 2) (206912_at), score: -0.47 FOXK2forkhead box K2 (203064_s_at), score: 0.53 FRMD4BFERM domain containing 4B (213056_at), score: 0.77 GABARAPL1GABA(A) receptor-associated protein like 1 (208868_s_at), score: -0.45 GABARAPL3GABA(A) receptors associated protein like 3 (pseudogene) (211458_s_at), score: -0.45 GABRR1gamma-aminobutyric acid (GABA) receptor, rho 1 (206525_at), score: 0.54 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (208217_at), score: -0.52 GAKcyclin G associated kinase (202281_at), score: 0.4 GASTgastrin (208138_at), score: -0.46 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (203178_at), score: 0.35 GEMIN8gem (nuclear organelle) associated protein 8 (219252_s_at), score: 0.47 GGTLC1gamma-glutamyltransferase light chain 1 (211416_x_at), score: -0.48 GLMNglomulin, FKBP associated protein (207153_s_at), score: -0.52 GLRA1glycine receptor, alpha 1 (207972_at), score: 0.51 GNA12guanine nucleotide binding protein (G protein) alpha 12 (221737_at), score: -0.57 GNL3LPguanine nucleotide binding protein-like 3 (nucleolar)-like pseudogene (220716_at), score: -0.61 GOLGA9Pgolgi autoantigen, golgin subfamily a, 9 pseudogene (213737_x_at), score: -0.45 GPATCH2G patch domain containing 2 (219078_at), score: -0.43 GPATCH8G patch domain containing 8 (212485_at), score: -0.45 GPR137BG protein-coupled receptor 137B (204137_at), score: -0.44 GPR144G protein-coupled receptor 144 (216289_at), score: -0.47 GPR4G protein-coupled receptor 4 (206236_at), score: 0.37 GPR63G protein-coupled receptor 63 (220993_s_at), score: 0.42 GPR65G protein-coupled receptor 65 (214467_at), score: 0.38 GPRASP1G protein-coupled receptor associated sorting protein 1 (204793_at), score: -0.52 GPX3glutathione peroxidase 3 (plasma) (214091_s_at), score: 0.58 GSTA1glutathione S-transferase alpha 1 (215766_at), score: -0.44 GSTCDglutathione S-transferase, C-terminal domain containing (220063_at), score: 0.81 GSTM4glutathione S-transferase mu 4 (204149_s_at), score: -0.46 GSTZ1glutathione transferase zeta 1 (209531_at), score: -0.5 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.46 GTF2H3general transcription factor IIH, polypeptide 3, 34kDa (222104_x_at), score: -0.57 GYG2glycogenin 2 (215695_s_at), score: -0.59 HAB1B1 for mucin (215778_x_at), score: -0.57 HAMPhepcidin antimicrobial peptide (220491_at), score: -0.55 HAND1heart and neural crest derivatives expressed 1 (220138_at), score: -0.65 HAVCR1hepatitis A virus cellular receptor 1 (207052_at), score: 0.38 HCG8HLA complex group 8 (215985_at), score: 0.89 HCLS1hematopoietic cell-specific Lyn substrate 1 (202957_at), score: -0.48 HDAC4histone deacetylase 4 (204225_at), score: -0.45 HINFPhistone H4 transcription factor (206495_s_at), score: -0.52 HIST1H2ALhistone cluster 1, H2al (214554_at), score: 0.41 HLA-DRB1major histocompatibility complex, class II, DR beta 1 (209312_x_at), score: 0.5 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.39 HOXA6homeobox A6 (208557_at), score: -0.47 HOXA7homeobox A7 (206848_at), score: 0.51 HOXB5homeobox B5 (205601_s_at), score: -0.45 HOXB6homeobox B6 (205366_s_at), score: 0.52 HPCAL4hippocalcin like 4 (219671_at), score: 0.61 HRASLSHRAS-like suppressor (219983_at), score: 0.55 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.45 HSF2heat shock transcription factor 2 (209657_s_at), score: -0.48 HYMAIhydatidiform mole associated and imprinted (non-protein coding) (215513_at), score: 0.69 ICA1islet cell autoantigen 1, 69kDa (211740_at), score: 0.7 IER3IP1immediate early response 3 interacting protein 1 (211406_at), score: -0.48 IFNGR1interferon gamma receptor 1 (211676_s_at), score: -0.54 IFT140intraflagellar transport 140 homolog (Chlamydomonas) (204792_s_at), score: -0.69 IGFALSinsulin-like growth factor binding protein, acid labile subunit (215712_s_at), score: -0.47 IGHG1immunoglobulin heavy constant gamma 1 (G1m marker) (217039_x_at), score: -0.72 IGLL3immunoglobulin lambda-like polypeptide 3 (215946_x_at), score: -0.44 IGSF3immunoglobulin superfamily, member 3 (202421_at), score: -0.47 IKZF4IKAROS family zinc finger 4 (Eos) (208472_at), score: -0.57 IMPACTImpact homolog (mouse) (218637_at), score: -0.51 INE1inactivation escape 1 (non-protein coding) (207252_at), score: -0.52 ING4inhibitor of growth family, member 4 (48825_at), score: -0.46 IQCGIQ motif containing G (221185_s_at), score: 0.42 IRF6interferon regulatory factor 6 (202597_at), score: 0.43 IRS1insulin receptor substrate 1 (204686_at), score: -0.48 ISOC1isochorismatase domain containing 1 (218170_at), score: -0.62 ITGA10integrin, alpha 10 (206766_at), score: -0.6 ITPKCinositol 1,4,5-trisphosphate 3-kinase C (213076_at), score: -0.49 JAK2Janus kinase 2 (a protein tyrosine kinase) (205842_s_at), score: 0.54 JMJD1Bjumonji domain containing 1B (210878_s_at), score: -0.55 JMJD1Cjumonji domain containing 1C (221763_at), score: -0.55 JTV1JTV1 gene (209972_s_at), score: -0.45 KBTBD4kelch repeat and BTB (POZ) domain containing 4 (218569_s_at), score: -0.7 KCND2potassium voltage-gated channel, Shal-related subfamily, member 2 (207103_at), score: 0.51 KCNIP1Kv channel interacting protein 1 (221307_at), score: 0.56 KCNJ14potassium inwardly-rectifying channel, subfamily J, member 14 (220776_at), score: 0.37 KCNJ16potassium inwardly-rectifying channel, subfamily J, member 16 (219564_at), score: 0.55 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (219287_at), score: -0.49 KIAA0562KIAA0562 (204075_s_at), score: -0.5 KIAA0892KIAA0892 (212505_s_at), score: 0.5 KIAA1107KIAA1107 (214098_at), score: 0.56 KIF26Bkinesin family member 26B (220002_at), score: -0.61 KLF1Kruppel-like factor 1 (erythroid) (210504_at), score: -0.51 KLF12Kruppel-like factor 12 (208467_at), score: 0.57 KLF7Kruppel-like factor 7 (ubiquitous) (204334_at), score: -0.51 KLHL3kelch-like 3 (Drosophila) (221221_s_at), score: 0.54 KRI1KRI1 homolog (S. cerevisiae) (218798_at), score: -0.45 KRT24keratin 24 (220267_at), score: 0.35 LAIR2leukocyte-associated immunoglobulin-like receptor 2 (207509_s_at), score: 0.42 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.46 LAT2linker for activation of T cells family, member 2 (211768_at), score: -0.61 LEPRleptin receptor (209894_at), score: -0.45 LGMNlegumain (201212_at), score: -0.49 LILRB3leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 3 (211133_x_at), score: -0.57 LILRP2leukocyte immunoglobulin-like receptor pseudogene 2 (214561_at), score: 0.57 LIMD1LIM domains containing 1 (218850_s_at), score: 0.37 LMF2lipase maturation factor 2 (212682_s_at), score: 0.39 LOC100129500hypothetical protein LOC100129500 (212884_x_at), score: -0.5 LOC100130703similar to hCG2042168 (207596_at), score: 0.55 LOC100130886hypothetical protein LOC100130886 (220486_x_at), score: -0.46 LOC100132247similar to Uncharacterized protein KIAA0220 (215002_at), score: 0.87 LOC100133105hypothetical protein LOC100133105 (214237_x_at), score: -0.51 LOC100133748similar to GTF2IRD2 protein (215569_at), score: -0.51 LOC100134713hypothetical protein LOC100134713 (222090_at), score: -0.68 LOC149501similar to keratin 8 (216821_at), score: -0.55 LOC257152hypothetical protein LOC257152 (215302_at), score: 0.5 LOC440354PI-3-kinase-related kinase SMG-1 pseudogene (210396_s_at), score: -0.53 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.36 LOC729806similar to hCG1725380 (217544_at), score: 0.7 LOC93432maltase-glucoamylase-like pseudogene (216666_at), score: 0.57 LRP3low density lipoprotein receptor-related protein 3 (204381_at), score: -0.47 LUC7LLUC7-like (S. cerevisiae) (220143_x_at), score: -0.47 LY6G5Clymphocyte antigen 6 complex, locus G5C (219860_at), score: 0.42 LYPLA2P1lysophospholipase II pseudogene 1 (216606_x_at), score: -0.44 MAN2B1mannosidase, alpha, class 2B, member 1 (209166_s_at), score: 0.38 MANBAmannosidase, beta A, lysosomal (203778_at), score: -0.73 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.35 MAP3K9mitogen-activated protein kinase kinase kinase 9 (213927_at), score: 0.74 MARCH8membrane-associated ring finger (C3HC4) 8 (221824_s_at), score: -0.44 MASP1mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) (213749_at), score: -0.47 MBIPMAP3K12 binding inhibitory protein 1 (218411_s_at), score: -0.47 MCM3APASMCM3AP antisense RNA (non-protein coding) (220459_at), score: 0.67 MFSD6major facilitator superfamily domain containing 6 (219858_s_at), score: 0.47 MGC5590hypothetical protein MGC5590 (220931_at), score: 0.35 MMP9matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) (203936_s_at), score: -0.52 MRPS6mitochondrial ribosomal protein S6 (213167_s_at), score: 0.49 MTMR3myotubularin related protein 3 (202197_at), score: 0.46 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.39 MUC3Amucin 3A, cell surface associated (217117_x_at), score: -0.54 MVKmevalonate kinase (36907_at), score: -0.46 MYBPC1myosin binding protein C, slow type (214087_s_at), score: 0.47 MYO15Amyosin XVA (220288_at), score: -0.47 MYO1Amyosin IA (211916_s_at), score: 0.49 MYO5Cmyosin VC (218966_at), score: 0.67 N4BP3Nedd4 binding protein 3 (214775_at), score: 0.37 NAIPNLR family, apoptosis inhibitory protein (204860_s_at), score: 0.42 NAT9N-acetyltransferase 9 (GCN5-related, putative) (204382_at), score: -0.5 NCOA1nuclear receptor coactivator 1 (209106_at), score: -0.59 NECAB3N-terminal EF-hand calcium binding protein 3 (210720_s_at), score: -0.44 NEURLneuralized homolog (Drosophila) (204889_s_at), score: -0.43 NFRKBnuclear factor related to kappaB binding protein (213028_at), score: -0.65 NFYCnuclear transcription factor Y, gamma (202215_s_at), score: 0.44 NLGN4Xneuroligin 4, X-linked (221933_at), score: 0.53 NMUneuromedin U (206023_at), score: 0.68 NOL12nucleolar protein 12 (219324_at), score: -0.45 NOTCH4Notch homolog 4 (Drosophila) (205247_at), score: -0.5 NPIPL2nuclear pore complex interacting protein-like 2 (221992_at), score: -0.51 NRCAMneuronal cell adhesion molecule (204105_s_at), score: 0.44 NSBP1nucleosomal binding protein 1 (221606_s_at), score: 0.43 NTRK2neurotrophic tyrosine kinase, receptor, type 2 (207152_at), score: 0.37 NXPH3neurexophilin 3 (221991_at), score: -0.44 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.47 OBFC2Boligonucleotide/oligosaccharide-binding fold containing 2B (218903_s_at), score: 0.39 OCEL1occludin/ELL domain containing 1 (205441_at), score: -0.49 OPLAH5-oxoprolinase (ATP-hydrolysing) (222025_s_at), score: -0.51 OR10C1olfactory receptor, family 10, subfamily C, member 1 (221339_at), score: 0.38 OR2S2olfactory receptor, family 2, subfamily S, member 2 (221409_at), score: 0.63 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.63 ORC1Lorigin recognition complex, subunit 1-like (yeast) (205085_at), score: 0.42 PAK4p21 protein (Cdc42/Rac)-activated kinase 4 (203154_s_at), score: 0.35 PAN2PAN2 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) (203117_s_at), score: -0.47 PAOXpolyamine oxidase (exo-N4-amino) (50400_at), score: -0.5 PARD6Apar-6 partitioning defective 6 homolog alpha (C. elegans) (205245_at), score: -0.57 PAX8paired box 8 (121_at), score: -0.51 PCCApropionyl Coenzyme A carboxylase, alpha polypeptide (203860_at), score: -0.48 PCDH11Yprotocadherin 11 Y-linked (211227_s_at), score: 0.97 PCIF1PDX1 C-terminal inhibiting factor 1 (222045_s_at), score: 0.38 PDCLphosducin-like (204449_at), score: -0.45 PDE3Aphosphodiesterase 3A, cGMP-inhibited (206388_at), score: 0.72 PDZRN4PDZ domain containing ring finger 4 (220595_at), score: 0.4 PERLD1per1-like domain containing 1 (55616_at), score: -0.46 PEX5peroxisomal biogenesis factor 5 (203244_at), score: -0.51 PGCprogastricsin (pepsinogen C) (205261_at), score: -0.5 PGLYRP4peptidoglycan recognition protein 4 (220944_at), score: -0.46 PGPEP1pyroglutamyl-peptidase I (219891_at), score: -0.55 PIAS4protein inhibitor of activated STAT, 4 (212881_at), score: -0.45 PIGNphosphatidylinositol glycan anchor biosynthesis, class N (219048_at), score: 0.38 PKLRpyruvate kinase, liver and RBC (222078_at), score: -0.56 PLAG1pleiomorphic adenoma gene 1 (205372_at), score: -0.44 PLEKHA9pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 9 (220157_x_at), score: -0.67 PLS1plastin 1 (I isoform) (205190_at), score: 0.65 PLXNB3plexin B3 (205957_at), score: 0.57 PMS2CLPMS2 C-terminal like pseudogene (221206_at), score: -0.5 PMS2L2postmeiotic segregation increased 2-like 2 pseudogene (215410_at), score: -0.44 POU6F1POU class 6 homeobox 1 (205878_at), score: -0.5 PPIAL4Apeptidylprolyl isomerase A (cyclophilin A)-like 4A (217136_at), score: 0.58 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (216347_s_at), score: -0.43 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: 0.37 PRDM10PR domain containing 10 (219515_at), score: -0.45 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (214474_at), score: -0.55 PRLprolactin (205445_at), score: 0.86 PROL1proline rich, lacrimal 1 (208004_at), score: 0.43 PSD4pleckstrin and Sec7 domain containing 4 (203317_at), score: -0.65 PTCD1pentatricopeptide repeat domain 1 (218956_s_at), score: -0.48 R3HDM2R3H domain containing 2 (203831_at), score: -0.63 RAB26RAB26, member RAS oncogene family (50965_at), score: -0.58 RAB30RAB30, member RAS oncogene family (206530_at), score: -0.62 RAB40BRAB40B, member RAS oncogene family (217597_x_at), score: -0.7 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.57 RAPGEF2Rap guanine nucleotide exchange factor (GEF) 2 (203097_s_at), score: -0.45 RASIP1Ras interacting protein 1 (220027_s_at), score: -0.57 RBM15RNA binding motif protein 15 (219286_s_at), score: -0.54 RCOR3REST corepressor 3 (218344_s_at), score: -0.46 RECQL5RecQ protein-like 5 (34063_at), score: 0.62 RIN1Ras and Rab interactor 1 (205211_s_at), score: 0.39 RIPK1receptor (TNFRSF)-interacting serine-threonine kinase 1 (209941_at), score: 0.39 RNASE2ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) (206111_at), score: 0.43 RP11-35N6.1plasticity related gene 3 (219732_at), score: 0.48 RPAINRPA interacting protein (216961_s_at), score: -0.47 RPAP1RNA polymerase II associated protein 1 (218441_s_at), score: 0.42 RPIAribose 5-phosphate isomerase A (212973_at), score: -0.59 RPL10P16ribosomal protein L10 pseudogene 16 (217680_x_at), score: -0.51 RUFY3RUN and FYVE domain containing 3 (210251_s_at), score: -0.48 SAA4serum amyloid A4, constitutive (207096_at), score: -0.56 SAMD14sterile alpha motif domain containing 14 (213866_at), score: -0.46 SCAMP5secretory carrier membrane protein 5 (212699_at), score: -0.52 SCARF1scavenger receptor class F, member 1 (206995_x_at), score: 0.49 SCDstearoyl-CoA desaturase (delta-9-desaturase) (200832_s_at), score: -0.43 SCN1Bsodium channel, voltage-gated, type I, beta (205508_at), score: -0.65 SEPT5septin 5 (209767_s_at), score: -0.45 SERGEFsecretion regulating guanine nucleotide exchange factor (220482_s_at), score: -0.6 SERPINA7serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 7 (206386_at), score: -0.45 SF4splicing factor 4 (215004_s_at), score: -0.64 SFNstratifin (33322_i_at), score: -0.52 SFXN1sideroflexin 1 (218392_x_at), score: -0.48 SH2D3ASH2 domain containing 3A (222169_x_at), score: -0.55 SIGLEC1sialic acid binding Ig-like lectin 1, sialoadhesin (44673_at), score: -0.64 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: 0.43 SIRT6sirtuin (silent mating type information regulation 2 homolog) 6 (S. cerevisiae) (219613_s_at), score: 0.35 SLC12A3solute carrier family 12 (sodium/chloride transporters), member 3 (215274_at), score: 0.42 SLC12A7solute carrier family 12 (potassium/chloride transporters), member 7 (218066_at), score: -0.43 SLC12A9solute carrier family 12 (potassium/chloride transporters), member 9 (220371_s_at), score: -0.47 SLC16A6solute carrier family 16, member 6 (monocarboxylic acid transporter 7) (207038_at), score: 0.62 SLC23A2solute carrier family 23 (nucleobase transporters), member 2 (209237_s_at), score: -0.46 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (218749_s_at), score: 0.4 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (211754_s_at), score: -0.52 SLC2A3solute carrier family 2 (facilitated glucose transporter), member 3 (202499_s_at), score: -0.47 SLC30A4solute carrier family 30 (zinc transporter), member 4 (207362_at), score: 0.5 SLC35C2solute carrier family 35, member C2 (219447_s_at), score: -0.55 SLC43A1solute carrier family 43, member 1 (204394_at), score: -0.54 SLC4A10solute carrier family 4, sodium bicarbonate transporter, member 10 (206830_at), score: 0.42 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.5 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (219795_at), score: 0.68 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (201349_at), score: -0.54 SMA5glucuronidase, beta pseudogene (215043_s_at), score: -0.45 SNORA21small nucleolar RNA, H/ACA box 21 (215224_at), score: -0.49 SP2Sp2 transcription factor (204367_at), score: -0.47 SPAG16sperm associated antigen 16 (219109_at), score: -0.53 SPAM1sperm adhesion molecule 1 (PH-20 hyaluronidase, zona pellucida binding) (216989_at), score: 0.54 SPATA2Lspermatogenesis associated 2-like (214965_at), score: -0.55 SPON1spondin 1, extracellular matrix protein (209436_at), score: 0.42 SPTA1spectrin, alpha, erythrocytic 1 (elliptocytosis 2) (206937_at), score: 0.91 SRBD1S1 RNA binding domain 1 (219055_at), score: -0.48 SRGAP2SLIT-ROBO Rho GTPase activating protein 2 (213329_at), score: -0.45 SSX3synovial sarcoma, X breakpoint 3 (211731_x_at), score: -0.53 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (220979_s_at), score: -0.44 ST7suppression of tumorigenicity 7 (207871_s_at), score: -0.47 ST8SIA3ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 3 (208064_s_at), score: -0.44 STRN4striatin, calmodulin binding protein 4 (217903_at), score: 0.36 STXBP5Lsyntaxin binding protein 5-like (215518_at), score: 0.35 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (212894_at), score: -0.66 SUV39H1suppressor of variegation 3-9 homolog 1 (Drosophila) (218619_s_at), score: 0.36 SYCP2synaptonemal complex protein 2 (206546_at), score: 0.56 TAAR2trace amine associated receptor 2 (221394_at), score: 0.75 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (202840_at), score: -0.46 TAF4BTAF4b RNA polymerase II, TATA box binding protein (TBP)-associated factor, 105kDa (216226_at), score: 0.35 TAPBPLTAP binding protein-like (218746_at), score: -0.56 TGFBR1transforming growth factor, beta receptor 1 (206943_at), score: -0.52 THADAthyroid adenoma associated (54632_at), score: -0.47 THOC6THO complex 6 homolog (Drosophila) (218848_at), score: -0.55 THUMPD1THUMP domain containing 1 (206555_s_at), score: -0.46 TIGD1Ltigger transposable element derived 1-like (216459_x_at), score: -0.44 TLR1toll-like receptor 1 (210176_at), score: 0.66 TMEM100transmembrane protein 100 (219230_at), score: 0.42 TMEM209transmembrane protein 209 (222267_at), score: -0.52 TNFRSF1Btumor necrosis factor receptor superfamily, member 1B (203508_at), score: -0.44 TOMM34translocase of outer mitochondrial membrane 34 (201870_at), score: 0.35 TRAPPC2P1trafficking protein particle complex 2 pseudogene 1 (209751_s_at), score: -0.47 TRAPPC9trafficking protein particle complex 9 (221836_s_at), score: -0.62 TRIM13tripartite motif-containing 13 (203659_s_at), score: -0.53 TRIM26tripartite motif-containing 26 (202702_at), score: 0.38 TRIM34tripartite motif-containing 34 (221044_s_at), score: 0.52 TRIM45tripartite motif-containing 45 (219923_at), score: -0.69 TRMT61BtRNA methyltransferase 61 homolog B (S. cerevisiae) (221229_s_at), score: -0.45 TROtrophinin (211700_s_at), score: -0.5 TRPM2transient receptor potential cation channel, subfamily M, member 2 (205708_s_at), score: -0.47 TSPAN31tetraspanin 31 (203227_s_at), score: -0.44 TSPY1testis specific protein, Y-linked 1 (216374_at), score: 0.43 TTBK2tau tubulin kinase 2 (213922_at), score: -0.46 TTC12tetratricopeptide repeat domain 12 (219587_at), score: -0.49 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.49 TUSC4tumor suppressor candidate 4 (203246_s_at), score: -0.47 TWISTNBTWIST neighbor (214729_at), score: 0.59 TXNthioredoxin (216609_at), score: -0.5 UBE2D1ubiquitin-conjugating enzyme E2D 1 (UBC4/5 homolog, yeast) (211764_s_at), score: -0.54 UBE4Bubiquitination factor E4B (UFD2 homolog, yeast) (202317_s_at), score: -0.44 USP6NLUSP6 N-terminal like (204761_at), score: -0.58 VASH1vasohibin 1 (203940_s_at), score: -0.47 VLDLRvery low density lipoprotein receptor (209822_s_at), score: -0.46 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (44111_at), score: -0.45 WDR48WD repeat domain 48 (222157_s_at), score: -0.43 WDR55WD repeat domain 55 (219809_at), score: 0.38 WDR60WD repeat domain 60 (219251_s_at), score: -0.46 WFDC8WAP four-disulfide core domain 8 (215276_at), score: -0.44 WNT2Bwingless-type MMTV integration site family, member 2B (206459_s_at), score: -0.43 WSB1WD repeat and SOCS box-containing 1 (201294_s_at), score: -0.46 WWC1WW and C2 domain containing 1 (213085_s_at), score: -0.55 WWOXWW domain containing oxidoreductase (219077_s_at), score: -0.46 YY2YY2 transcription factor (216531_at), score: 0.42 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.59 ZBTB40zinc finger and BTB domain containing 40 (203958_s_at), score: -0.6 ZBTB6zinc finger and BTB domain containing 6 (206098_at), score: 0.36 ZCCHC4zinc finger, CCHC domain containing 4 (220473_s_at), score: 0.46 ZCCHC6zinc finger, CCHC domain containing 6 (220933_s_at), score: -0.53 ZDHHC14zinc finger, DHHC-type containing 14 (219247_s_at), score: -0.52 ZFP30zinc finger protein 30 homolog (mouse) (207090_x_at), score: -0.49 ZKSCAN4zinc finger with KRAB and SCAN domains 4 (213625_at), score: -0.46 ZKSCAN5zinc finger with KRAB and SCAN domains 5 (203730_s_at), score: 0.52 ZNF133zinc finger protein 133 (216960_s_at), score: -0.56 ZNF14zinc finger protein 14 (219854_at), score: -0.48 ZNF174zinc finger protein 174 (210290_at), score: -0.47 ZNF175zinc finger protein 175 (205497_at), score: 0.58 ZNF253zinc finger protein 253 (206900_x_at), score: -0.49 ZNF287zinc finger protein 287 (216710_x_at), score: 0.55 ZNF329zinc finger protein 329 (219765_at), score: -0.56 ZNF334zinc finger protein 334 (220022_at), score: 0.35 ZNF426zinc finger protein 426 (205964_at), score: -0.57 ZNF529zinc finger protein 529 (215307_at), score: -0.6 ZNF592zinc finger protein 592 (204473_s_at), score: -0.44 ZNF614zinc finger protein 614 (220721_at), score: 0.44 ZNF652zinc finger protein 652 (205594_at), score: -0.55 ZNF669zinc finger protein 669 (220215_at), score: -0.48 ZNF749zinc finger protein 749 (215289_at), score: 0.44 ZNF783zinc finger family member 783 (221876_at), score: -0.51 ZNF839zinc finger protein 839 (221709_s_at), score: -0.51 ZNF91zinc finger protein 91 (206059_at), score: -0.49 ZNHIT6zinc finger, HIT type 6 (218932_at), score: -0.44
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486171.cel | 27 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485911.cel | 14 | 6 | Cycle | hgu133a2 | none | Cycle 1 |