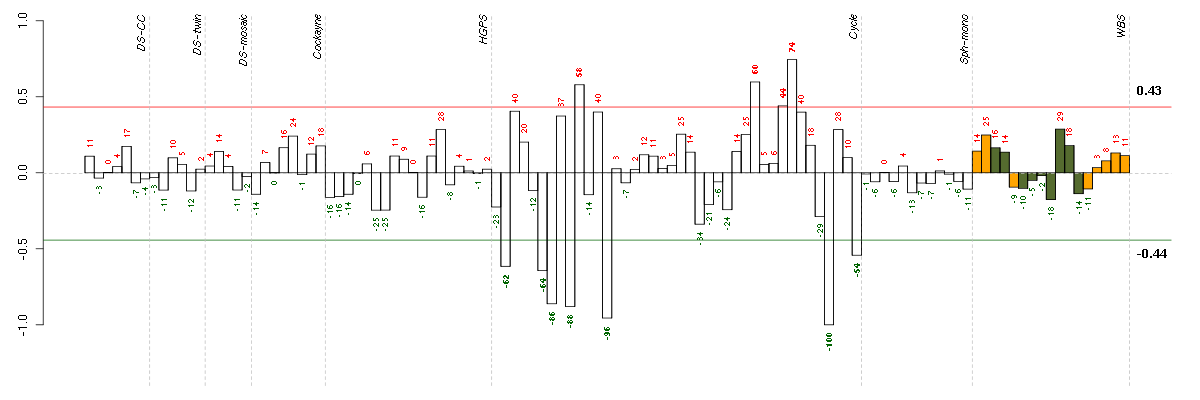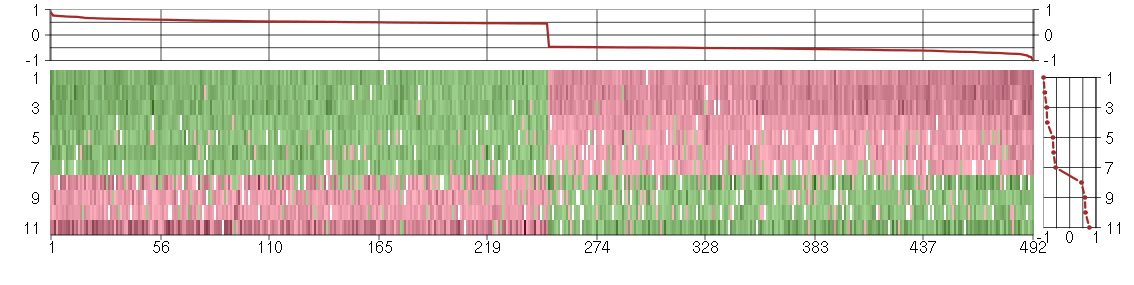

Under-expression is coded with green,
over-expression with red color.

digestion
The whole of the physical, chemical, and biochemical processes carried out by multicellular organisms to break down ingested nutrients into components that may be easily absorbed and directed into metabolism.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
positive regulation of catalytic activity
Any process that activates or increases the activity of an enzyme.
regulation of GTPase activity
Any process that modulates the rate of GTP hydrolysis by a GTPase.
positive regulation of GTPase activity
Any process that activates or increases the activity of a GTPase.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of hydrolase activity
Any process that modulates the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
positive regulation of hydrolase activity
Any process that activates or increases the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of molecular functions. Molecular functions are elemental biological activities occurring at the molecular level, such as catalysis or binding.
all
This term is the most general term possible
positive regulation of hydrolase activity
Any process that activates or increases the frequency, rate or extent of hydrolase activity, the catalysis of the hydrolysis of various bonds.
positive regulation of GTPase activity
Any process that activates or increases the activity of a GTPase.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
This term is the most general term possible
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

calcium ion binding
Interacting selectively with calcium ions (Ca2+).
molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively with any metal ion.
ion binding
Interacting selectively with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively with cations, charged atoms or groups of atoms with a net positive charge.
all
This term is the most general term possible
calcium ion binding
Interacting selectively with calcium ions (Ca2+).
ABCA8ATP-binding cassette, sub-family A (ABC1), member 8 (204719_at), score: 0.47 ABCC5ATP-binding cassette, sub-family C (CFTR/MRP), member 5 (209380_s_at), score: -0.5 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.52 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: -0.49 ABHD3abhydrolase domain containing 3 (213017_at), score: -0.51 ABHD5abhydrolase domain containing 5 (218739_at), score: -0.52 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.48 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.54 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.67 ACSS3acyl-CoA synthetase short-chain family member 3 (219616_at), score: 0.54 ACTC1actin, alpha, cardiac muscle 1 (205132_at), score: -0.54 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.69 ADAM19ADAM metallopeptidase domain 19 (meltrin beta) (209765_at), score: -0.47 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.69 ADH1Balcohol dehydrogenase 1B (class I), beta polypeptide (209612_s_at), score: 0.45 ADORA2Badenosine A2b receptor (205891_at), score: 0.72 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.5 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.47 AGRNagrin (217419_x_at), score: 0.47 AHI1Abelson helper integration site 1 (221569_at), score: -0.7 AIM1absent in melanoma 1 (212543_at), score: 0.66 AJAP1adherens junctions associated protein 1 (206460_at), score: -0.47 AKAP2A kinase (PRKA) anchor protein 2 (202759_s_at), score: 0.47 AKR1B10aldo-keto reductase family 1, member B10 (aldose reductase) (206561_s_at), score: 0.57 AKR1C3aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II) (209160_at), score: 0.54 ANGPTL2angiopoietin-like 2 (213001_at), score: 0.62 ANXA10annexin A10 (210143_at), score: -0.8 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.52 APOBEC3Fapolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3F (214995_s_at), score: 0.49 APOL6apolipoprotein L, 6 (219716_at), score: 0.63 ARAP1ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 1 (34206_at), score: 0.46 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.55 ARHGEF2rho/rac guanine nucleotide exchange factor (GEF) 2 (209435_s_at), score: 0.6 ARL4CADP-ribosylation factor-like 4C (202207_at), score: 0.47 ARMC9armadillo repeat containing 9 (219637_at), score: 0.47 ARSJarylsulfatase family, member J (219973_at), score: -0.52 ASAP3ArfGAP with SH3 domain, ankyrin repeat and PH domain 3 (219103_at), score: 0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.63 ATP6V0BATPase, H+ transporting, lysosomal 21kDa, V0 subunit b (200078_s_at), score: -0.49 ATP6V1B2ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B2 (201089_at), score: 0.47 B9D1B9 protein domain 1 (210534_s_at), score: 0.48 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.46 BAZ1Abromodomain adjacent to zinc finger domain, 1A (217985_s_at), score: -0.48 BCAS3breast carcinoma amplified sequence 3 (220488_s_at), score: 0.5 BDKRB2bradykinin receptor B2 (205870_at), score: 0.53 BIN1bridging integrator 1 (210201_x_at), score: 0.61 BMP6bone morphogenetic protein 6 (206176_at), score: -0.58 BPHLbiphenyl hydrolase-like (serine hydrolase) (205750_at), score: 0.46 BRIP1BRCA1 interacting protein C-terminal helicase 1 (221703_at), score: -0.48 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.61 BTG2BTG family, member 2 (201236_s_at), score: 0.62 C10orf26chromosome 10 open reading frame 26 (202808_at), score: 0.48 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.59 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.53 C13orf15chromosome 13 open reading frame 15 (218723_s_at), score: 0.63 C14orf147chromosome 14 open reading frame 147 (212460_at), score: -0.48 C14orf159chromosome 14 open reading frame 159 (218298_s_at), score: 0.45 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.49 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.49 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.48 C1orf107chromosome 1 open reading frame 107 (214193_s_at), score: -0.54 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.49 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.71 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.57 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.5 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.51 CALB2calbindin 2 (205428_s_at), score: -0.62 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.72 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.6 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.47 CASZ1castor zinc finger 1 (220015_at), score: -0.49 CCDC28Acoiled-coil domain containing 28A (209479_at), score: 0.51 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.47 CCNE2cyclin E2 (205034_at), score: -0.51 CCNG1cyclin G1 (208796_s_at), score: 0.5 CCNT2cyclin T2 (204645_at), score: -0.51 CD248CD248 molecule, endosialin (219025_at), score: 0.48 CD55CD55 molecule, decay accelerating factor for complement (Cromer blood group) (201925_s_at), score: -0.47 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.57 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.52 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.56 CDCP1CUB domain containing protein 1 (218451_at), score: -0.49 CDH13cadherin 13, H-cadherin (heart) (204726_at), score: -0.49 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.65 CDKN1Ccyclin-dependent kinase inhibitor 1C (p57, Kip2) (213348_at), score: 0.58 CEP68centrosomal protein 68kDa (212677_s_at), score: 0.49 CHMP5chromatin modifying protein 5 (218085_at), score: -0.47 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.54 CHRNA10cholinergic receptor, nicotinic, alpha 10 (220210_at), score: -0.49 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.66 CICcapicua homolog (Drosophila) (212784_at), score: 0.54 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.5 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.66 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.6 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.51 COL18A1collagen, type XVIII, alpha 1 (209081_s_at), score: 0.46 CRBNcereblon (218142_s_at), score: 0.52 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.88 CScitrate synthase (208660_at), score: 0.46 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.56 CTDSP2CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 (203445_s_at), score: 0.54 CUL7cullin 7 (36084_at), score: 0.49 CUX1cut-like homeobox 1 (214743_at), score: -0.51 DAPK1death-associated protein kinase 1 (203139_at), score: 0.52 DCKdeoxycytidine kinase (203302_at), score: -0.46 DCLK1doublecortin-like kinase 1 (205399_at), score: 0.59 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.52 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.56 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.53 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (212834_at), score: -0.48 DEDDdeath effector domain containing (215158_s_at), score: 0.48 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.62 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.55 DEPDC6DEP domain containing 6 (218858_at), score: 0.64 DERL2Der1-like domain family, member 2 (218333_at), score: -0.52 DHRS3dehydrogenase/reductase (SDR family) member 3 (202481_at), score: 0.64 DIRAS3DIRAS family, GTP-binding RAS-like 3 (215506_s_at), score: -0.57 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.66 DMWDdystrophia myotonica, WD repeat containing (213231_at), score: 0.45 DMXL2Dmx-like 2 (212820_at), score: -0.47 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.77 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.47 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.56 DNM1dynamin 1 (215116_s_at), score: 0.56 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.65 DOPEY1dopey family member 1 (40612_at), score: -0.47 DRAMdamage-regulated autophagy modulator (218627_at), score: 0.45 DTX4deltex homolog 4 (Drosophila) (212611_at), score: 0.58 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.49 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.7 DZIP3DAZ interacting protein 3, zinc finger (213186_at), score: 0.53 EAF2ELL associated factor 2 (219551_at), score: -0.62 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.48 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.61 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.61 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.46 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.47 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.49 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.7 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.6 ELK3ELK3, ETS-domain protein (SRF accessory protein 2) (221773_at), score: -0.49 ELTD1EGF, latrophilin and seven transmembrane domain containing 1 (219134_at), score: -0.55 ENOX1ecto-NOX disulfide-thiol exchanger 1 (219501_at), score: 0.53 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.56 EPHA4EPH receptor A4 (206114_at), score: -0.6 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (202017_at), score: 0.47 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.67 ESRRAestrogen-related receptor alpha (1487_at), score: -0.49 FAM102Afamily with sequence similarity 102, member A (212400_at), score: 0.47 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.52 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.61 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.48 FARSAphenylalanyl-tRNA synthetase, alpha subunit (216602_s_at), score: -0.49 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.63 FERMT1fermitin family homolog 1 (Drosophila) (218796_at), score: -0.52 FGGYFGGY carbohydrate kinase domain containing (219718_at), score: 0.54 FICDFIC domain containing (219910_at), score: -0.57 FLJ11151hypothetical protein FLJ11151 (218610_s_at), score: -0.47 FLJ12529pre-mRNA cleavage factor I, 59 kDa subunit (217866_at), score: 0.47 FLJ40113golgi autoantigen, golgin subfamily a-like pseudogene (213212_x_at), score: -0.5 FLOT1flotillin 1 (210142_x_at), score: 0.46 FNBP1formin binding protein 1 (212288_at), score: 0.61 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.56 FOLR3folate receptor 3 (gamma) (206371_at), score: -0.53 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.46 FOXK2forkhead box K2 (203064_s_at), score: 0.6 FRG1FSHD region gene 1 (204145_at), score: -0.52 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.51 FYNFYN oncogene related to SRC, FGR, YES (210105_s_at), score: 0.49 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.59 GABBR2gamma-aminobutyric acid (GABA) B receptor, 2 (209990_s_at), score: -0.52 GALgalanin prepropeptide (214240_at), score: -0.52 GALK2galactokinase 2 (205219_s_at), score: -0.54 GALNT12UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) (218885_s_at), score: 0.55 GAS7growth arrest-specific 7 (202191_s_at), score: 0.48 GDF15growth differentiation factor 15 (221577_x_at), score: 0.52 GDF5growth differentiation factor 5 (206614_at), score: 0.48 GDPD5glycerophosphodiester phosphodiesterase domain containing 5 (32502_at), score: 0.53 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -0.72 GHRgrowth hormone receptor (205498_at), score: -0.56 GKglycerol kinase (207387_s_at), score: -0.68 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.79 GLTSCR2glioma tumor suppressor candidate region gene 2 (217807_s_at), score: 0.5 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.72 GMPRguanosine monophosphate reductase (204187_at), score: 0.57 GNG11guanine nucleotide binding protein (G protein), gamma 11 (204115_at), score: -0.47 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.64 GPM6Bglycoprotein M6B (209170_s_at), score: -0.55 GPR177G protein-coupled receptor 177 (221958_s_at), score: -0.61 GPR65G protein-coupled receptor 65 (214467_at), score: 0.46 GPRC5BG protein-coupled receptor, family C, group 5, member B (203632_s_at), score: 0.5 GRAMD1BGRAM domain containing 1B (212906_at), score: -0.51 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: -0.54 GSK3Bglycogen synthase kinase 3 beta (209945_s_at), score: 0.49 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.74 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.5 GUF1GUF1 GTPase homolog (S. cerevisiae) (218884_s_at), score: -0.49 H1FXH1 histone family, member X (204805_s_at), score: 0.53 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.57 HHIPL2HHIP-like 2 (220283_at), score: -0.52 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.69 HLA-DQB1major histocompatibility complex, class II, DQ beta 1 (211654_x_at), score: -0.47 HNRNPDheterogeneous nuclear ribonucleoprotein D (AU-rich element RNA binding protein 1, 37kDa) (221480_at), score: -0.49 HOXC5homeobox C5 (206739_at), score: -0.5 HPNhepsin (204934_s_at), score: -0.48 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.71 HSD17B7hydroxysteroid (17-beta) dehydrogenase 7 (220081_x_at), score: 0.54 HSPA12Aheat shock 70kDa protein 12A (214434_at), score: 0.69 HSPA4Lheat shock 70kDa protein 4-like (205543_at), score: 0.46 ICAM3intercellular adhesion molecule 3 (204949_at), score: -0.48 ID4inhibitor of DNA binding 4, dominant negative helix-loop-helix protein (209291_at), score: -0.55 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.5 IFIH1interferon induced with helicase C domain 1 (219209_at), score: 0.57 IL13RA2interleukin 13 receptor, alpha 2 (206172_at), score: -0.51 IL33interleukin 33 (209821_at), score: -0.66 IRF1interferon regulatory factor 1 (202531_at), score: 0.57 IRF2interferon regulatory factor 2 (203275_at), score: 0.45 IRS1insulin receptor substrate 1 (204686_at), score: -0.53 ITGA1integrin, alpha 1 (214660_at), score: -0.53 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.57 ITPR2inositol 1,4,5-triphosphate receptor, type 2 (202660_at), score: -0.47 JUNDjun D proto-oncogene (203751_x_at), score: 0.6 JUPjunction plakoglobin (201015_s_at), score: 0.59 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.85 KCNJ8potassium inwardly-rectifying channel, subfamily J, member 8 (205303_at), score: -0.56 KIAA0562KIAA0562 (204075_s_at), score: -0.51 KIAA1305KIAA1305 (220911_s_at), score: 0.76 KIAA1462KIAA1462 (213316_at), score: 0.63 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.74 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.66 LDLRlow density lipoprotein receptor (202068_s_at), score: 0.6 LEPREL1leprecan-like 1 (218717_s_at), score: -0.48 LHFPL2lipoma HMGIC fusion partner-like 2 (212658_at), score: 0.52 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.47 LIN7Blin-7 homolog B (C. elegans) (219760_at), score: -0.55 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.71 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.49 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.73 LOC391132similar to hCG2041276 (216177_at), score: -0.51 LOC399491LOC399491 protein (214035_x_at), score: 0.65 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.58 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.48 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.5 LRIG1leucine-rich repeats and immunoglobulin-like domains 1 (211596_s_at), score: 0.47 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.76 LRRC40leucine rich repeat containing 40 (218577_at), score: -0.49 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.65 LYPD1LY6/PLAUR domain containing 1 (212909_at), score: -0.61 MAN1C1mannosidase, alpha, class 1C, member 1 (218918_at), score: 0.72 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.55 MBPmyelin basic protein (210136_at), score: 0.45 MC4Rmelanocortin 4 receptor (221467_at), score: -0.74 MDM2Mdm2 p53 binding protein homolog (mouse) (205386_s_at), score: 0.52 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.66 MGC87042similar to Six transmembrane epithelial antigen of prostate (217553_at), score: -0.53 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.6 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.68 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.69 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.6 MTUS1mitochondrial tumor suppressor 1 (212096_s_at), score: 0.52 MX2myxovirus (influenza virus) resistance 2 (mouse) (204994_at), score: 0.56 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.63 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.48 NCALDneurocalcin delta (211685_s_at), score: -0.6 NCKAP1NCK-associated protein 1 (217465_at), score: -0.48 NDRG4NDRG family member 4 (209159_s_at), score: 0.55 NDUFA2NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 2, 8kDa (213550_s_at), score: 0.52 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (213012_at), score: -0.54 NEFLneurofilament, light polypeptide (221805_at), score: -0.57 NEFMneurofilament, medium polypeptide (205113_at), score: -1 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.51 NEO1neogenin homolog 1 (chicken) (204321_at), score: 0.52 NETO2neuropilin (NRP) and tolloid (TLL)-like 2 (218888_s_at), score: -0.52 NF1neurofibromin 1 (211094_s_at), score: 0.55 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.48 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.52 NINJ1ninjurin 1 (203045_at), score: 0.47 NMIN-myc (and STAT) interactor (203964_at), score: 0.53 NOX4NADPH oxidase 4 (219773_at), score: -0.52 NPC2Niemann-Pick disease, type C2 (200701_at), score: 0.54 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.74 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.53 NPTX1neuronal pentraxin I (204684_at), score: -0.73 NR1H3nuclear receptor subfamily 1, group H, member 3 (203920_at), score: 0.56 NR5A2nuclear receptor subfamily 5, group A, member 2 (208343_s_at), score: -0.47 NRXN3neurexin 3 (205795_at), score: -0.58 NTMneurotrimin (222020_s_at), score: -0.55 NUDT11nudix (nucleoside diphosphate linked moiety X)-type motif 11 (219855_at), score: 0.48 NUP160nucleoporin 160kDa (212709_at), score: -0.65 NUPL2nucleoporin like 2 (204003_s_at), score: -0.5 OAS12',5'-oligoadenylate synthetase 1, 40/46kDa (205552_s_at), score: 0.5 OAS22'-5'-oligoadenylate synthetase 2, 69/71kDa (204972_at), score: 0.66 OAS32'-5'-oligoadenylate synthetase 3, 100kDa (218400_at), score: 0.53 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.58 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.55 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.52 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.74 ORAI3ORAI calcium release-activated calcium modulator 3 (221864_at), score: 0.61 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.7 P2RX5purinergic receptor P2X, ligand-gated ion channel, 5 (210448_s_at), score: -0.59 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.73 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.6 PCDH9protocadherin 9 (219737_s_at), score: -0.86 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.45 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.48 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.67 PDCD1LG2programmed cell death 1 ligand 2 (220049_s_at), score: -0.62 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.6 PDE7Bphosphodiesterase 7B (220343_at), score: -0.51 PDE8Aphosphodiesterase 8A (212522_at), score: -0.49 PDGFRAplatelet-derived growth factor receptor, alpha polypeptide (203131_at), score: 0.45 PECRperoxisomal trans-2-enoyl-CoA reductase (221142_s_at), score: 0.53 PFN2profilin 2 (204992_s_at), score: 0.51 PI4K2Aphosphatidylinositol 4-kinase type 2 alpha (209345_s_at), score: 0.46 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (213889_at), score: 0.56 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.65 PKN1protein kinase N1 (202161_at), score: 0.49 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (210145_at), score: -0.49 PLA2G4Cphospholipase A2, group IVC (cytosolic, calcium-independent) (209785_s_at), score: 0.48 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.48 PLCD1phospholipase C, delta 1 (205125_at), score: 0.51 PLEKHA5pleckstrin homology domain containing, family A member 5 (220952_s_at), score: 0.48 PMP22peripheral myelin protein 22 (210139_s_at), score: 0.51 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.63 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.45 POM121L2POM121 membrane glycoprotein-like 2 (rat) (216582_at), score: -0.51 POMZP3POM (POM121 homolog, rat) and ZP3 fusion (204148_s_at), score: 0.47 PORP450 (cytochrome) oxidoreductase (208928_at), score: 0.51 PPARGperoxisome proliferator-activated receptor gamma (208510_s_at), score: 0.61 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.56 PPP2R1Bprotein phosphatase 2 (formerly 2A), regulatory subunit A, beta isoform (202884_s_at), score: -0.49 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.51 PRKCHprotein kinase C, eta (218764_at), score: 0.45 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.6 PRPS2phosphoribosyl pyrophosphate synthetase 2 (203401_at), score: -0.51 PRR5proline rich 5 (renal) (47069_at), score: 0.5 PRSS1protease, serine, 1 (trypsin 1) (216470_x_at), score: -0.5 PRSS2protease, serine, 2 (trypsin 2) (205402_x_at), score: -0.56 PRSS3protease, serine, 3 (207463_x_at), score: -0.51 PSMB8proteasome (prosome, macropain) subunit, beta type, 8 (large multifunctional peptidase 7) (209040_s_at), score: 0.52 PTGS1prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) (215813_s_at), score: -0.46 PTP4A1protein tyrosine phosphatase type IVA, member 1 (200730_s_at), score: -0.55 PTPN9protein tyrosine phosphatase, non-receptor type 9 (202958_at), score: 0.45 PTPROprotein tyrosine phosphatase, receptor type, O (211600_at), score: 0.61 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.56 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.47 RAB11FIP1RAB11 family interacting protein 1 (class I) (219681_s_at), score: 0.45 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.65 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.66 RANBP6RAN binding protein 6 (213019_at), score: -0.56 RASSF2Ras association (RalGDS/AF-6) domain family member 2 (203185_at), score: 0.56 RBM15BRNA binding motif protein 15B (202689_at), score: 0.51 REEP1receptor accessory protein 1 (204364_s_at), score: -0.52 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.75 RFX5regulatory factor X, 5 (influences HLA class II expression) (202963_at), score: 0.49 RHBDF1rhomboid 5 homolog 1 (Drosophila) (218686_s_at), score: 0.53 RINT1RAD50 interactor 1 (218598_at), score: -0.49 RNF11ring finger protein 11 (208924_at), score: -0.58 RNF138ring finger protein 138 (218738_s_at), score: -0.5 RNF187ring finger protein 187 (212155_at), score: 0.5 RNF220ring finger protein 220 (219988_s_at), score: 0.64 RNF44ring finger protein 44 (203286_at), score: 0.52 RNFT1ring finger protein, transmembrane 1 (221194_s_at), score: -0.47 ROR1receptor tyrosine kinase-like orphan receptor 1 (205805_s_at), score: 0.47 ROR2receptor tyrosine kinase-like orphan receptor 2 (205578_at), score: 0.58 RP11-345P4.4similar to solute carrier family 35, member E2 (217122_s_at), score: 0.49 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.52 RP4-691N24.1ninein-like (207705_s_at), score: 0.46 RPL13Aribosomal protein L13a (200715_x_at), score: 0.46 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.61 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.52 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.61 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.59 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.54 RSC1A1regulatory solute carrier protein, family 1, member 1 (214583_at), score: -0.46 S1PR1sphingosine-1-phosphate receptor 1 (204642_at), score: -0.53 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: -0.53 SAP30LSAP30-like (219129_s_at), score: 0.49 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.64 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.5 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.62 SDF2L1stromal cell-derived factor 2-like 1 (218681_s_at), score: -0.62 SECTM1secreted and transmembrane 1 (213716_s_at), score: 0.49 SELENBP1selenium binding protein 1 (214433_s_at), score: 0.46 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (204614_at), score: -0.59 SERPINB9serpin peptidase inhibitor, clade B (ovalbumin), member 9 (209723_at), score: 0.55 SESN1sestrin 1 (218346_s_at), score: 0.57 SF1splicing factor 1 (208313_s_at), score: 0.53 SGSHN-sulfoglucosamine sulfohydrolase (35626_at), score: 0.57 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.49 SH3D19SH3 domain containing 19 (211620_x_at), score: 0.5 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.47 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.56 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.49 SIRT3sirtuin (silent mating type information regulation 2 homolog) 3 (S. cerevisiae) (221562_s_at), score: 0.52 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.68 SLC2A6solute carrier family 2 (facilitated glucose transporter), member 6 (220091_at), score: 0.58 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.6 SLC35A3solute carrier family 35 (UDP-N-acetylglucosamine (UDP-GlcNAc) transporter), member A3 (206770_s_at), score: -0.48 SLC39A7solute carrier family 39 (zinc transporter), member 7 (202667_s_at), score: -0.59 SLC4A4solute carrier family 4, sodium bicarbonate cotransporter, member 4 (203908_at), score: -0.55 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -0.7 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (207053_at), score: -0.52 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.66 SMAD3SMAD family member 3 (218284_at), score: 0.62 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.6 SNCAsynuclein, alpha (non A4 component of amyloid precursor) (204466_s_at), score: 0.46 SNNstannin (218032_at), score: 0.6 SNX27sorting nexin family member 27 (221006_s_at), score: 0.51 SOAT1sterol O-acyltransferase 1 (221561_at), score: -0.54 SPATA20spermatogenesis associated 20 (218164_at), score: 0.52 SPRY1sprouty homolog 1, antagonist of FGF signaling (Drosophila) (212558_at), score: 0.47 SSTR1somatostatin receptor 1 (208482_at), score: -0.62 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.48 ST5suppression of tumorigenicity 5 (202440_s_at), score: 0.53 STARD5StAR-related lipid transfer (START) domain containing 5 (213820_s_at), score: 0.54 STAT6signal transducer and activator of transcription 6, interleukin-4 induced (201331_s_at), score: 0.47 STAU1staufen, RNA binding protein, homolog 1 (Drosophila) (207320_x_at), score: 0.46 STIM1stromal interaction molecule 1 (202764_at), score: 0.59 SV2Asynaptic vesicle glycoprotein 2A (203069_at), score: 0.5 SYNE1spectrin repeat containing, nuclear envelope 1 (209447_at), score: -0.49 SYNJ1synaptojanin 1 (212990_at), score: -0.61 SYNPOsynaptopodin (202796_at), score: 0.55 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.56 TAPBPTAP binding protein (tapasin) (208829_at), score: 0.53 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.65 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (204526_s_at), score: 0.6 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.6 TCERG1transcription elongation regulator 1 (202396_at), score: -0.49 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.64 TFDP2transcription factor Dp-2 (E2F dimerization partner 2) (203588_s_at), score: 0.52 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.58 THSD7Athrombospondin, type I, domain containing 7A (214920_at), score: 0.6 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.73 TLR3toll-like receptor 3 (206271_at), score: 0.63 TM4SF1transmembrane 4 L six family member 1 (209386_at), score: -0.52 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.47 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.55 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.59 TMEM140transmembrane protein 140 (218999_at), score: 0.53 TMEM2transmembrane protein 2 (218113_at), score: -0.66 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.58 TMOD1tropomodulin 1 (203661_s_at), score: -0.47 TMSB15Bthymosin beta 15B (214051_at), score: 0.49 TNFAIP2tumor necrosis factor, alpha-induced protein 2 (202510_s_at), score: 0.53 TNFAIP6tumor necrosis factor, alpha-induced protein 6 (206026_s_at), score: 0.61 TNFRSF10Ctumor necrosis factor receptor superfamily, member 10c, decoy without an intracellular domain (206222_at), score: 0.5 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.59 TNS3tensin 3 (217853_at), score: 0.53 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.51 TOP3Atopoisomerase (DNA) III alpha (214300_s_at), score: -0.51 TP53tumor protein p53 (201746_at), score: 0.76 TRIM21tripartite motif-containing 21 (204804_at), score: 0.75 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.64 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.55 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.63 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.5 TSKUtsukushin (218245_at), score: 0.58 TSPAN13tetraspanin 13 (217979_at), score: -0.56 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.51 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.57 TXLNAtaxilin alpha (212300_at), score: 0.48 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.58 UBE2L6ubiquitin-conjugating enzyme E2L 6 (201649_at), score: 0.5 UBXN1UBX domain protein 1 (201871_s_at), score: 0.5 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.47 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.54 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.67 ULBP2UL16 binding protein 2 (221291_at), score: -0.74 ULK1unc-51-like kinase 1 (C. elegans) (209333_at), score: 0.67 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.49 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.52 VWA5Avon Willebrand factor A domain containing 5A (210102_at), score: 0.62 WBP2WW domain binding protein 2 (209117_at), score: 0.59 WDR4WD repeat domain 4 (221632_s_at), score: -0.63 WDR42AWD repeat domain 42A (202249_s_at), score: 0.52 WDR6WD repeat domain 6 (217734_s_at), score: 0.7 XPCxeroderma pigmentosum, complementation group C (209375_at), score: 0.46 XYLBxylulokinase homolog (H. influenzae) (214776_x_at), score: -0.48 XYLT1xylosyltransferase I (213725_x_at), score: -0.5 YDD19YDD19 protein (37079_at), score: -0.49 YES1v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 (202932_at), score: 0.55 ZBTB20zinc finger and BTB domain containing 20 (205383_s_at), score: 0.46 ZBTB22zinc finger and BTB domain containing 22 (213081_at), score: 0.46 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.47 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.47 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.71 ZFP106zinc finger protein 106 homolog (mouse) (217781_s_at), score: 0.46 ZFP36L1zinc finger protein 36, C3H type-like 1 (211965_at), score: 0.46 ZNF358zinc finger protein 358 (219379_x_at), score: 0.46 ZNF580zinc finger protein 580 (220748_s_at), score: 0.53 ZNF706zinc finger protein 706 (218059_at), score: 0.53
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485671.cel | 2 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486431.cel | 40 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486271.cel | 32 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |