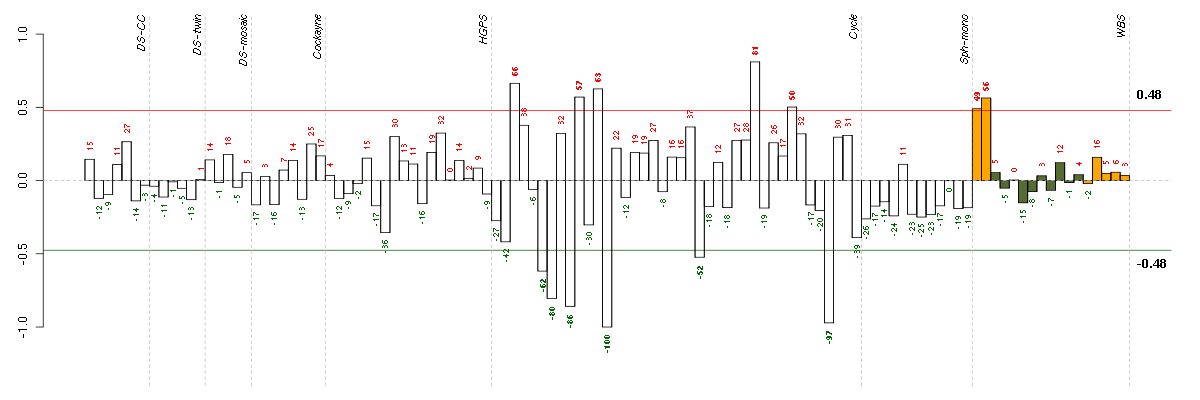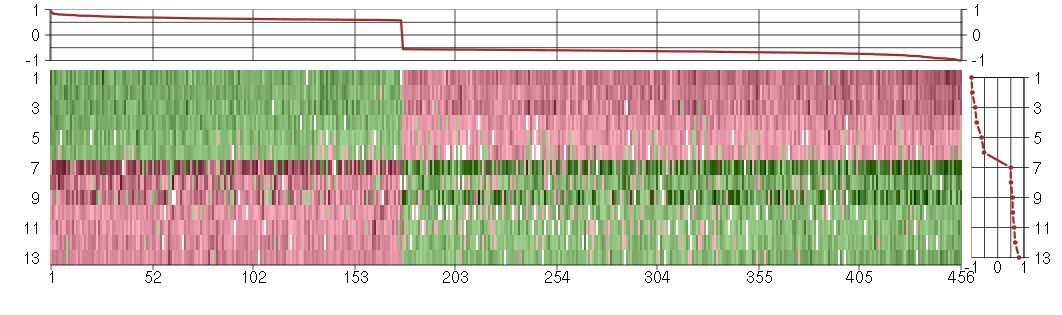

Under-expression is coded with green,
over-expression with red color.



AAASachalasia, adrenocortical insufficiency, alacrimia (Allgrove, triple-A) (218075_at), score: 0.65 ABCD1ATP-binding cassette, sub-family D (ALD), member 1 (205142_x_at), score: 0.7 ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (201873_s_at), score: -0.79 ABLIM1actin binding LIM protein 1 (200965_s_at), score: 0.59 ACADSBacyl-Coenzyme A dehydrogenase, short/branched chain (205355_at), score: -0.58 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (212476_at), score: -0.58 ACN9ACN9 homolog (S. cerevisiae) (218981_at), score: -0.61 ACSL1acyl-CoA synthetase long-chain family member 1 (201963_at), score: -0.7 ACSL4acyl-CoA synthetase long-chain family member 4 (202422_s_at), score: -0.93 ACTR3BARP3 actin-related protein 3 homolog B (yeast) (218868_at), score: -0.73 ADARadenosine deaminase, RNA-specific (201786_s_at), score: 0.64 ADCK2aarF domain containing kinase 2 (221893_s_at), score: 0.68 ADORA2Badenosine A2b receptor (205891_at), score: 0.7 AGBL5ATP/GTP binding protein-like 5 (218480_at), score: 0.66 AGPAT11-acylglycerol-3-phosphate O-acyltransferase 1 (lysophosphatidic acid acyltransferase, alpha) (215535_s_at), score: 0.73 AHI1Abelson helper integration site 1 (221569_at), score: -0.73 AIM1absent in melanoma 1 (212543_at), score: 0.61 ALDH1L1aldehyde dehydrogenase 1 family, member L1 (215798_at), score: -0.69 ANXA10annexin A10 (210143_at), score: -0.64 ANXA2P1annexin A2 pseudogene 1 (210876_at), score: -0.59 AP2A2adaptor-related protein complex 2, alpha 2 subunit (212159_x_at), score: 0.64 APAF1apoptotic peptidase activating factor 1 (204859_s_at), score: -0.67 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (202211_at), score: -0.64 ARSAarylsulfatase A (204443_at), score: 0.7 ATF2activating transcription factor 2 (212984_at), score: -0.61 ATF5activating transcription factor 5 (204999_s_at), score: 0.62 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (211137_s_at), score: -0.72 ATP8B1ATPase, class I, type 8B, member 1 (214594_x_at), score: -0.74 AXLAXL receptor tyrosine kinase (202685_s_at), score: 0.59 AZI25-azacytidine induced 2 (218043_s_at), score: -0.58 B9D1B9 protein domain 1 (210534_s_at), score: 0.63 BAP1BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) (201419_at), score: 0.6 BAT2HLA-B associated transcript 2 (212081_x_at), score: 0.72 BAT3HLA-B associated transcript 3 (213318_s_at), score: 0.65 BBS10Bardet-Biedl syndrome 10 (219487_at), score: -0.56 BCL2L1BCL2-like 1 (215037_s_at), score: 0.59 BMI1BMI1 polycomb ring finger oncogene (202265_at), score: -0.59 BMP6bone morphogenetic protein 6 (206176_at), score: -0.59 BNIP2BCL2/adenovirus E1B 19kDa interacting protein 2 (209308_s_at), score: -0.59 BRWD1bromodomain and WD repeat domain containing 1 (214820_at), score: -0.57 BTBD2BTB (POZ) domain containing 2 (207722_s_at), score: 0.83 C10orf97chromosome 10 open reading frame 97 (218297_at), score: -0.63 C12orf29chromosome 12 open reading frame 29 (213701_at), score: -0.65 C14orf45chromosome 14 open reading frame 45 (220173_at), score: -0.6 C14orf94chromosome 14 open reading frame 94 (218383_at), score: 0.75 C15orf33chromosome 15 open reading frame 33 (216411_s_at), score: -0.64 C16orf45chromosome 16 open reading frame 45 (212736_at), score: 0.62 C1orf109chromosome 1 open reading frame 109 (218712_at), score: -0.63 C1orf9chromosome 1 open reading frame 9 (203429_s_at), score: -0.68 C22orf30chromosome 22 open reading frame 30 (216555_at), score: -0.57 C3orf18chromosome 3 open reading frame 18 (219114_at), score: 0.63 C3orf64chromosome 3 open reading frame 64 (221935_s_at), score: -0.63 C4orf43chromosome 4 open reading frame 43 (218513_at), score: -0.95 C5orf44chromosome 5 open reading frame 44 (218674_at), score: -0.91 C6orf211chromosome 6 open reading frame 211 (218195_at), score: -0.64 C7orf64chromosome 7 open reading frame 64 (221596_s_at), score: -0.68 CA9carbonic anhydrase IX (205199_at), score: 0.65 CABIN1calcineurin binding protein 1 (37652_at), score: 0.63 CALB2calbindin 2 (205428_s_at), score: -0.78 CALCOCO1calcium binding and coiled-coil domain 1 (209002_s_at), score: 0.81 CAPN5calpain 5 (205166_at), score: 0.63 CAPZA2capping protein (actin filament) muscle Z-line, alpha 2 (201237_at), score: -0.69 CASC3cancer susceptibility candidate 3 (207842_s_at), score: 0.6 CASP7caspase 7, apoptosis-related cysteine peptidase (207181_s_at), score: -0.57 CCDC41coiled-coil domain containing 41 (219644_at), score: -0.67 CCDC76coiled-coil domain containing 76 (219130_at), score: -0.61 CCNE2cyclin E2 (205034_at), score: -0.59 CCNT2cyclin T2 (204645_at), score: -0.58 CD248CD248 molecule, endosialin (219025_at), score: 0.64 CD2APCD2-associated protein (203593_at), score: -0.62 CDC25Bcell division cycle 25 homolog B (S. pombe) (201853_s_at), score: 0.62 CDC2L6cell division cycle 2-like 6 (CDK8-like) (212899_at), score: 0.64 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (217849_s_at), score: 0.68 CDC42EP4CDC42 effector protein (Rho GTPase binding) 4 (214721_x_at), score: 0.6 CDC73cell division cycle 73, Paf1/RNA polymerase II complex component, homolog (S. cerevisiae) (218578_at), score: -0.64 CDH6cadherin 6, type 2, K-cadherin (fetal kidney) (205532_s_at), score: -0.64 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (203341_at), score: -0.64 CHMP5chromatin modifying protein 5 (218085_at), score: -0.71 CHORDC1cysteine and histidine-rich domain (CHORD)-containing 1 (218566_s_at), score: -0.82 CHUKconserved helix-loop-helix ubiquitous kinase (209666_s_at), score: -0.86 CICcapicua homolog (Drosophila) (212784_at), score: 0.82 CIZ1CDKN1A interacting zinc finger protein 1 (205516_x_at), score: 0.76 CLIP3CAP-GLY domain containing linker protein 3 (212358_at), score: 0.66 CMPK1cytidine monophosphate (UMP-CMP) kinase 1, cytosolic (217870_s_at), score: -0.77 CNOT3CCR4-NOT transcription complex, subunit 3 (203239_s_at), score: 0.75 CRCPCGRP receptor component (203898_at), score: -0.68 CRTC3CREB regulated transcription coactivator 3 (218648_at), score: 0.92 CSGALNACT2chondroitin sulfate N-acetylgalactosaminyltransferase 2 (222235_s_at), score: -0.68 CSNK2A2casein kinase 2, alpha prime polypeptide (203575_at), score: 0.61 CTSEcathepsin E (205927_s_at), score: -0.57 CUL7cullin 7 (36084_at), score: 0.68 CYP3A4cytochrome P450, family 3, subfamily A, polypeptide 4 (205998_x_at), score: -0.76 DAXXdeath-domain associated protein (201763_s_at), score: 0.63 DCP2DCP2 decapping enzyme homolog (S. cerevisiae) (212919_at), score: -0.77 DDAH2dimethylarginine dimethylaminohydrolase 2 (215537_x_at), score: 0.7 DDB2damage-specific DNA binding protein 2, 48kDa (203409_at), score: 0.65 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (212834_at), score: -0.61 DENND2ADENN/MADD domain containing 2A (53991_at), score: 0.65 DENND4CDENN/MADD domain containing 4C (205684_s_at), score: -0.58 DENND5BDENN/MADD domain containing 5B (215058_at), score: -0.72 DERL2Der1-like domain family, member 2 (218333_at), score: -0.58 DKFZP564O0823DKFZP564O0823 protein (204687_at), score: -0.69 DMPKdystrophia myotonica-protein kinase (37996_s_at), score: 0.69 DNAJB9DnaJ (Hsp40) homolog, subfamily B, member 9 (202843_at), score: -0.91 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (213853_at), score: -0.56 DNAJC6DnaJ (Hsp40) homolog, subfamily C, member 6 (204720_s_at), score: -0.69 DNPEPaspartyl aminopeptidase (201937_s_at), score: 0.58 DOCK10dedicator of cytokinesis 10 (219279_at), score: -0.63 DOCK9dedicator of cytokinesis 9 (212538_at), score: -0.62 DOPEY1dopey family member 1 (40612_at), score: -0.82 DUSP14dual specificity phosphatase 14 (203367_at), score: -0.58 DUSP4dual specificity phosphatase 4 (204014_at), score: -0.63 EAF2ELL associated factor 2 (219551_at), score: -0.82 ECM1extracellular matrix protein 1 (209365_s_at), score: 0.69 EDEM1ER degradation enhancer, mannosidase alpha-like 1 (203279_at), score: -0.6 EDNRAendothelin receptor type A (204464_s_at), score: -0.57 EEA1early endosome antigen 1 (204840_s_at), score: -0.63 EGFepidermal growth factor (beta-urogastrone) (206254_at), score: -0.65 EHBP1L1EH domain binding protein 1-like 1 (221755_at), score: 0.63 EHMT2euchromatic histone-lysine N-methyltransferase 2 (202326_at), score: 0.58 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (218696_at), score: -0.61 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (201143_s_at), score: -0.59 EIF3Meukaryotic translation initiation factor 3, subunit M (202231_at), score: -0.6 EIF4Eeukaryotic translation initiation factor 4E (201436_at), score: -0.92 EIF4G3eukaryotic translation initiation factor 4 gamma, 3 (201936_s_at), score: -0.56 EIF5Beukaryotic translation initiation factor 5B (214314_s_at), score: -0.74 ELK3ELK3, ETS-domain protein (SRF accessory protein 2) (221773_at), score: -0.56 EMCNendomucin (219436_s_at), score: 0.59 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (209473_at), score: -0.79 EPHA4EPH receptor A4 (206114_at), score: -0.71 EPS15epidermal growth factor receptor pathway substrate 15 (217887_s_at), score: -0.58 ERBB2IPerbb2 interacting protein (217941_s_at), score: -0.66 ESM1endothelial cell-specific molecule 1 (208394_x_at), score: -0.56 EVI1ecotropic viral integration site 1 (221884_at), score: -0.57 EVI5ecotropic viral integration site 5 (209717_at), score: -0.56 FAM155Afamily with sequence similarity 155, member A (214825_at), score: -0.61 FAM18Bfamily with sequence similarity 18, member B (218446_s_at), score: -0.92 FAM29Afamily with sequence similarity 29, member A (218602_s_at), score: -0.57 FAM3Cfamily with sequence similarity 3, member C (201889_at), score: -0.62 FAM49Bfamily with sequence similarity 49, member B (217916_s_at), score: -0.56 FBXL14F-box and leucine-rich repeat protein 14 (213145_at), score: 0.61 FCER1GFc fragment of IgE, high affinity I, receptor for; gamma polypeptide (204232_at), score: -0.58 FKBP8FK506 binding protein 8, 38kDa (208255_s_at), score: 0.58 FLOT1flotillin 1 (210142_x_at), score: 0.68 FNBP1formin binding protein 1 (212288_at), score: 0.68 FNDC3Afibronectin type III domain containing 3A (202304_at), score: -0.59 FOXC2forkhead box C2 (MFH-1, mesenchyme forkhead 1) (214520_at), score: 0.67 FPGTfucose-1-phosphate guanylyltransferase (205140_at), score: -0.63 FRG1FSHD region gene 1 (204145_at), score: -0.7 FUBP3far upstream element (FUSE) binding protein 3 (212824_at), score: -0.74 G3BP2GTPase activating protein (SH3 domain) binding protein 2 (208841_s_at), score: -0.77 GFOD1glucose-fructose oxidoreductase domain containing 1 (219821_s_at), score: -1 GGA3golgi associated, gamma adaptin ear containing, ARF binding protein 3 (209411_s_at), score: 0.58 GHRgrowth hormone receptor (205498_at), score: -0.68 GKglycerol kinase (207387_s_at), score: -0.67 GK3Pglycerol kinase 3 pseudogene (215966_x_at), score: -0.79 GMCL1germ cell-less homolog 1 (Drosophila) (218458_at), score: -0.76 GNPTABN-acetylglucosamine-1-phosphate transferase, alpha and beta subunits (212959_s_at), score: -0.6 GOLGA8Ggolgi autoantigen, golgin subfamily a, 8G (222149_x_at), score: -0.61 GOLT1Bgolgi transport 1 homolog B (S. cerevisiae) (218193_s_at), score: -0.69 GP1BBglycoprotein Ib (platelet), beta polypeptide (206655_s_at), score: 0.58 GPM6Bglycoprotein M6B (209170_s_at), score: -0.61 GRSF1G-rich RNA sequence binding factor 1 (201501_s_at), score: -0.68 GSK3Bglycogen synthase kinase 3 beta (209945_s_at), score: 0.6 GTF2A1Lgeneral transcription factor IIA, 1-like (213413_at), score: 0.64 GTF2F2general transcription factor IIF, polypeptide 2, 30kDa (209595_at), score: -0.65 H1F0H1 histone family, member 0 (208886_at), score: 0.59 H1FXH1 histone family, member X (204805_s_at), score: 0.71 HBS1LHBS1-like (S. cerevisiae) (209314_s_at), score: -0.68 HDAC5histone deacetylase 5 (202455_at), score: 0.59 HDAC6histone deacetylase 6 (206846_s_at), score: 0.61 HDGFhepatoma-derived growth factor (high-mobility group protein 1-like) (200896_x_at), score: 0.69 HISPPD1histidine acid phosphatase domain containing 1 (203253_s_at), score: -0.87 HMG20Bhigh-mobility group 20B (210719_s_at), score: 0.67 HMGA1high mobility group AT-hook 1 (210457_x_at), score: 0.68 HNRNPH2heterogeneous nuclear ribonucleoprotein H2 (H') (201132_at), score: -0.73 HNRNPUL1heterogeneous nuclear ribonucleoprotein U-like 1 (209675_s_at), score: 0.62 HSD17B6hydroxysteroid (17-beta) dehydrogenase 6 homolog (mouse) (37512_at), score: -0.89 HSPA9heat shock 70kDa protein 9 (mortalin) (200690_at), score: -0.57 HUS1HUS1 checkpoint homolog (S. pombe) (217618_x_at), score: -0.68 IBTKinhibitor of Bruton agammaglobulinemia tyrosine kinase (210970_s_at), score: -0.67 IDUAiduronidase, alpha-L- (205059_s_at), score: 0.61 IFRD1interferon-related developmental regulator 1 (202146_at), score: -0.63 IGHMBP2immunoglobulin mu binding protein 2 (215980_s_at), score: -0.63 IL22RA1interleukin 22 receptor, alpha 1 (220056_at), score: -0.59 IMPACTImpact homolog (mouse) (218637_at), score: -0.59 INTS1integrator complex subunit 1 (212212_s_at), score: 0.62 IRF9interferon regulatory factor 9 (203882_at), score: 0.63 IRS1insulin receptor substrate 1 (204686_at), score: -0.57 ISOC1isochorismatase domain containing 1 (218170_at), score: -0.62 ITGA1integrin, alpha 1 (214660_at), score: -0.63 ITGAVintegrin, alpha V (vitronectin receptor, alpha polypeptide, antigen CD51) (202351_at), score: -0.67 JMJD1Cjumonji domain containing 1C (221763_at), score: -0.6 JMJD7jumonji domain containing 7 (60528_at), score: -0.66 JUNDjun D proto-oncogene (203751_x_at), score: 0.77 JUPjunction plakoglobin (201015_s_at), score: 0.74 KAL1Kallmann syndrome 1 sequence (205206_at), score: -0.95 KIAA0562KIAA0562 (204075_s_at), score: -0.75 KIAA1305KIAA1305 (220911_s_at), score: 0.67 KIAA1462KIAA1462 (213316_at), score: 0.6 KLHL23kelch-like 23 (Drosophila) (213610_s_at), score: -0.71 LARP4La ribonucleoprotein domain family, member 4 (214155_s_at), score: -0.94 LARSleucyl-tRNA synthetase (217810_x_at), score: -0.65 LIG4ligase IV, DNA, ATP-dependent (206235_at), score: -0.57 LLGL2lethal giant larvae homolog 2 (Drosophila) (203713_s_at), score: -0.66 LOC100132540similar to LOC339047 protein (214870_x_at), score: 0.74 LOC128192similar to peptidyl-Pro cis trans isomerase (217346_at), score: -0.68 LOC339047hypothetical protein LOC339047 (221501_x_at), score: 0.69 LOC391132similar to hCG2041276 (216177_at), score: -0.81 LOC399491LOC399491 protein (214035_x_at), score: 0.62 LOC440434hypothetical protein FLJ11822 (215090_x_at), score: 0.75 LPGAT1lysophosphatidylglycerol acyltransferase 1 (202651_at), score: -0.77 LRDDleucine-rich repeats and death domain containing (221640_s_at), score: 0.66 LRP8low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (205282_at), score: -0.84 LRRC40leucine rich repeat containing 40 (218577_at), score: -0.68 LRRN3leucine rich repeat neuronal 3 (209841_s_at), score: -0.64 MAGOH2mago-nashi homolog 2, proliferation-associated (Drosophila) (217693_x_at), score: -0.62 MAP1Smicrotubule-associated protein 1S (218522_s_at), score: 0.65 MAP2microtubule-associated protein 2 (210015_s_at), score: -0.56 MAP2K5mitogen-activated protein kinase kinase 5 (211370_s_at), score: 0.71 MAP3K6mitogen-activated protein kinase kinase kinase 6 (219278_at), score: 0.74 MAPK1IP1Lmitogen-activated protein kinase 1 interacting protein 1-like (212499_s_at), score: -0.67 MAPK3mitogen-activated protein kinase 3 (212046_x_at), score: 0.6 MAPK7mitogen-activated protein kinase 7 (35617_at), score: 0.61 MATR3matrin 3 (200626_s_at), score: -0.66 MC4Rmelanocortin 4 receptor (221467_at), score: -0.62 MED15mediator complex subunit 15 (222175_s_at), score: 0.58 MED23mediator complex subunit 23 (218846_at), score: -0.6 MEN1multiple endocrine neoplasia I (202645_s_at), score: 0.64 MEX3Cmex-3 homolog C (C. elegans) (218247_s_at), score: -0.59 MFAP3Lmicrofibrillar-associated protein 3-like (205442_at), score: -0.66 MINK1misshapen-like kinase 1 (zebrafish) (214246_x_at), score: 0.71 MIS12MIS12, MIND kinetochore complex component, homolog (yeast) (221559_s_at), score: -0.62 MMP10matrix metallopeptidase 10 (stromelysin 2) (205680_at), score: -0.73 MMP16matrix metallopeptidase 16 (membrane-inserted) (207012_at), score: -0.65 MPHOSPH10M-phase phosphoprotein 10 (U3 small nucleolar ribonucleoprotein) (212885_at), score: -0.57 MPP1membrane protein, palmitoylated 1, 55kDa (202974_at), score: 0.6 MSTO1misato homolog 1 (Drosophila) (218296_x_at), score: -0.6 MTMR11myotubularin related protein 11 (205076_s_at), score: 0.62 MUC1mucin 1, cell surface associated (207847_s_at), score: 0.76 MYBBP1AMYB binding protein (P160) 1a (219098_at), score: -0.59 MYL4myosin, light chain 4, alkali; atrial, embryonic (210088_x_at), score: -0.7 NARG1LNMDA receptor regulated 1-like (219378_at), score: -0.69 NAT13N-acetyltransferase 13 (GCN5-related) (217745_s_at), score: -0.69 NCALDneurocalcin delta (211685_s_at), score: -0.6 NCKAP1NCK-associated protein 1 (217465_at), score: -0.71 NCRNA00093non-protein coding RNA 93 (210723_x_at), score: -0.61 NDUFA5NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 5, 13kDa (201304_at), score: -0.67 NEFMneurofilament, medium polypeptide (205113_at), score: -0.88 NENFneuron derived neurotrophic factor (218407_x_at), score: 0.72 NF1neurofibromin 1 (211094_s_at), score: 0.65 NFATC4nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 4 (205897_at), score: 0.71 NFKBIAnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha (201502_s_at), score: 0.75 NHLRC2NHL repeat containing 2 (219353_at), score: -0.61 NOL10nucleolar protein 10 (218591_s_at), score: -0.59 NOX4NADPH oxidase 4 (219773_at), score: -0.7 NPIPnuclear pore complex interacting protein (204538_x_at), score: 0.83 NPR2natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) (204310_s_at), score: 0.69 NPTX1neuronal pentraxin I (204684_at), score: -0.57 NUP160nucleoporin 160kDa (212709_at), score: -0.75 NUPL2nucleoporin like 2 (204003_s_at), score: -0.6 OBFC2Aoligonucleotide/oligosaccharide-binding fold containing 2A (219334_s_at), score: -0.69 OLFML2Bolfactomedin-like 2B (213125_at), score: 0.67 OLR1oxidized low density lipoprotein (lectin-like) receptor 1 (210004_at), score: -0.79 OPA1optic atrophy 1 (autosomal dominant) (212213_x_at), score: -0.62 OPCMLopioid binding protein/cell adhesion molecule-like (214111_at), score: -0.69 OR7E156Polfactory receptor, family 7, subfamily E, member 156 pseudogene (222327_x_at), score: -0.59 OSTM1osteopetrosis associated transmembrane protein 1 (218196_at), score: -0.87 P2RY5purinergic receptor P2Y, G-protein coupled, 5 (218589_at), score: 0.64 PAK1p21 protein (Cdc42/Rac)-activated kinase 1 (209615_s_at), score: 0.65 PAQR3progestin and adipoQ receptor family member III (213372_at), score: -0.81 PARVBparvin, beta (204629_at), score: 0.69 PCDH9protocadherin 9 (219737_s_at), score: -0.94 PCDHG@protocadherin gamma cluster (215836_s_at), score: 0.61 PCDHGA1protocadherin gamma subfamily A, 1 (209079_x_at), score: 0.61 PCDHGA11protocadherin gamma subfamily A, 11 (211876_x_at), score: 0.7 PCDHGA3protocadherin gamma subfamily A, 3 (216352_x_at), score: 0.65 PCTK1PCTAIRE protein kinase 1 (208824_x_at), score: 0.61 PCYT2phosphate cytidylyltransferase 2, ethanolamine (209577_at), score: 0.64 PDCLphosducin-like (204449_at), score: -0.69 PDE5Aphosphodiesterase 5A, cGMP-specific (206757_at), score: 0.61 PEX6peroxisomal biogenesis factor 6 (204545_at), score: 0.74 PHF16PHD finger protein 16 (204866_at), score: -0.6 PIGOphosphatidylinositol glycan anchor biosynthesis, class O (209998_at), score: 0.61 PINK1PTEN induced putative kinase 1 (209018_s_at), score: 0.65 PIP5K1Cphosphatidylinositol-4-phosphate 5-kinase, type I, gamma (212518_at), score: 0.63 PITX1paired-like homeodomain 1 (208502_s_at), score: 0.65 PKN1protein kinase N1 (202161_at), score: 0.7 PLAUplasminogen activator, urokinase (211668_s_at), score: 0.6 PLCD1phospholipase C, delta 1 (205125_at), score: 0.76 PLD3phospholipase D family, member 3 (201050_at), score: 0.65 PNPLA2patatin-like phospholipase domain containing 2 (212705_x_at), score: 0.79 PNPLA6patatin-like phospholipase domain containing 6 (203718_at), score: 0.6 POLR2Apolymerase (RNA) II (DNA directed) polypeptide A, 220kDa (202725_at), score: 0.62 PPATphosphoribosyl pyrophosphate amidotransferase (209433_s_at), score: -0.72 PPFIA1protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 1 (202066_at), score: -0.56 PPP1CBprotein phosphatase 1, catalytic subunit, beta isoform (201407_s_at), score: -0.6 PPP3R1protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform (204507_s_at), score: -0.62 PRKCDprotein kinase C, delta (202545_at), score: 0.64 PRKD2protein kinase D2 (209282_at), score: 0.59 PRMT3protein arginine methyltransferase 3 (213320_at), score: -0.91 PTOV1prostate tumor overexpressed 1 (212032_s_at), score: 0.6 PVRL3poliovirus receptor-related 3 (213325_at), score: -0.74 PXNpaxillin (211823_s_at), score: 0.64 RAB11BRAB11B, member RAS oncogene family (34478_at), score: 0.65 RAB1ARAB1A, member RAS oncogene family (213440_at), score: -0.86 RAB33BRAB33B, member RAS oncogene family (221014_s_at), score: -0.63 RAB7ARAB7A, member RAS oncogene family (211960_s_at), score: -0.67 RABGGTBRab geranylgeranyltransferase, beta subunit (213704_at), score: -0.7 RANBP6RAN binding protein 6 (213019_at), score: -0.74 RBM15BRNA binding motif protein 15B (202689_at), score: 0.72 RECQLRecQ protein-like (DNA helicase Q1-like) (205091_x_at), score: -0.61 REREarginine-glutamic acid dipeptide (RE) repeats (200940_s_at), score: 0.78 RIF1RAP1 interacting factor homolog (yeast) (214700_x_at), score: -0.56 RNF11ring finger protein 11 (208924_at), score: -0.72 RNF138ring finger protein 138 (218738_s_at), score: -0.57 RNF220ring finger protein 220 (219988_s_at), score: 0.78 RNF44ring finger protein 44 (203286_at), score: 0.6 RP2retinitis pigmentosa 2 (X-linked recessive) (205191_at), score: -0.69 RP3-377H14.5hypothetical LOC285830 (222279_at), score: -0.57 RP5-1000E10.4suppressor of IKK epsilon (221705_s_at), score: -0.73 RP5-886K2.1neuronal thread protein AD7c-NTP (207953_at), score: -0.57 RPL18AP6ribosomal protein L18a pseudogene 6 (216383_at), score: -0.58 RPL21P68ribosomal protein L21 pseudogene 68 (217340_at), score: -0.6 RPLP2P1ribosomal protein, large P2, pseudogene 1 (216490_x_at), score: -0.68 RPS10P11ribosomal protein S10 pseudogene 11 (216505_x_at), score: -0.62 RPS17P5ribosomal protein S17 pseudogene 5 (216348_at), score: -0.82 RPS25ribosomal protein S25 (200091_s_at), score: -0.67 RPS27Aribosomal protein S27a (217144_at), score: -0.57 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (219037_at), score: -0.83 RRP1Bribosomal RNA processing 1 homolog B (S. cerevisiae) (212846_at), score: -0.61 RSC1A1regulatory solute carrier protein, family 1, member 1 (214583_at), score: -0.59 SACM1LSAC1 suppressor of actin mutations 1-like (yeast) (202797_at), score: -0.71 SAP30LSAP30-like (219129_s_at), score: 0.61 SASH1SAM and SH3 domain containing 1 (213236_at), score: 0.59 SAV1salvador homolog 1 (Drosophila) (218276_s_at), score: 0.71 SBNO2strawberry notch homolog 2 (Drosophila) (204166_at), score: 0.79 SCAMP4secretory carrier membrane protein 4 (213244_at), score: 0.68 SCAPSREBF chaperone (212329_at), score: 0.59 SDPRserum deprivation response (phosphatidylserine binding protein) (218711_s_at), score: 0.59 SEC62SEC62 homolog (S. cerevisiae) (208942_s_at), score: -0.64 SEP1515 kDa selenoprotein (200902_at), score: -0.56 SERPINB1serpin peptidase inhibitor, clade B (ovalbumin), member 1 (212268_at), score: 0.6 SFXN1sideroflexin 1 (218392_x_at), score: -0.57 SH3BP5SH3-domain binding protein 5 (BTK-associated) (201810_s_at), score: 0.61 SH3GLB2SH3-domain GRB2-like endophilin B2 (218813_s_at), score: 0.72 SIGIRRsingle immunoglobulin and toll-interleukin 1 receptor (TIR) domain (52940_at), score: 0.8 SIGLEC6sialic acid binding Ig-like lectin 6 (210796_x_at), score: -0.67 SIP1survival of motor neuron protein interacting protein 1 (205063_at), score: -0.59 SIX1SIX homeobox 1 (205817_at), score: 0.59 SLC25A13solute carrier family 25, member 13 (citrin) (203775_at), score: -0.78 SLC25A32solute carrier family 25, member 32 (221020_s_at), score: -0.65 SLC2A4RGSLC2A4 regulator (218494_s_at), score: 0.63 SLC30A9solute carrier family 30 (zinc transporter), member 9 (202614_at), score: -0.67 SLC33A1solute carrier family 33 (acetyl-CoA transporter), member 1 (203165_s_at), score: -0.71 SLC39A14solute carrier family 39 (zinc transporter), member 14 (212110_at), score: -0.58 SLC4A7solute carrier family 4, sodium bicarbonate cotransporter, member 7 (209884_s_at), score: -1 SLC5A5solute carrier family 5 (sodium iodide symporter), member 5 (211123_at), score: -0.64 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (203578_s_at), score: -0.59 SLMO2slowmo homolog 2 (Drosophila) (217851_s_at), score: -0.91 SMAD3SMAD family member 3 (218284_at), score: 0.66 SMCHD1structural maintenance of chromosomes flexible hinge domain containing 1 (212569_at), score: -0.64 SMG6Smg-6 homolog, nonsense mediated mRNA decay factor (C. elegans) (214940_s_at), score: 0.62 SMN1survival of motor neuron 1, telomeric (203852_s_at), score: -0.76 SNX27sorting nexin family member 27 (221006_s_at), score: 0.64 SOAT1sterol O-acyltransferase 1 (221561_at), score: -0.7 SOLHsmall optic lobes homolog (Drosophila) (204275_at), score: 0.61 SORBS3sorbin and SH3 domain containing 3 (209253_at), score: 0.63 SP3Sp3 transcription factor (213168_at), score: -0.67 SPATA20spermatogenesis associated 20 (218164_at), score: 0.65 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (213140_s_at), score: -0.57 SSTR1somatostatin receptor 1 (208482_at), score: -0.64 ST20suppressor of tumorigenicity 20 (210242_x_at), score: -0.66 STIM1stromal interaction molecule 1 (202764_at), score: 0.59 STRN3striatin, calmodulin binding protein 3 (204496_at), score: -0.58 STXBP3syntaxin binding protein 3 (203310_at), score: -0.57 SULT1B1sulfotransferase family, cytosolic, 1B, member 1 (207601_at), score: -0.69 SUPT4H1suppressor of Ty 4 homolog 1 (S. cerevisiae) (201484_at), score: 0.59 SYNJ1synaptojanin 1 (212990_at), score: -0.65 SYNPOsynaptopodin (202796_at), score: 0.61 TAF13TAF13 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 18kDa (205966_at), score: -0.62 TAF2TAF2 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 150kDa (209523_at), score: -0.78 TASP1taspase, threonine aspartase, 1 (219443_at), score: -0.6 TBC1D2BTBC1 domain family, member 2B (212796_s_at), score: 0.62 TBCCD1TBCC domain containing 1 (206451_at), score: -0.62 TCEB1transcription elongation factor B (SIII), polypeptide 1 (15kDa, elongin C) (202823_at), score: -0.7 TCERG1transcription elongation regulator 1 (202396_at), score: -0.56 TDGthymine-DNA glycosylase (203743_s_at), score: -0.64 TENC1tensin like C1 domain containing phosphatase (tensin 2) (212494_at), score: 0.79 TERF2telomeric repeat binding factor 2 (203611_at), score: 0.59 TGDSTDP-glucose 4,6-dehydratase (208249_s_at), score: -0.73 TIAF1TGFB1-induced anti-apoptotic factor 1 (202039_at), score: 0.78 TICAM2toll-like receptor adaptor molecule 2 (214658_at), score: -0.98 TIMP2TIMP metallopeptidase inhibitor 2 (203167_at), score: 0.6 TLE4transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) (204872_at), score: -0.63 TM9SF3transmembrane 9 superfamily member 3 (217758_s_at), score: -0.71 TMED7transmembrane emp24 protein transport domain containing 7 (209404_s_at), score: -0.6 TMEFF1transmembrane protein with EGF-like and two follistatin-like domains 1 (205122_at), score: -0.6 TMEM123transmembrane protein 123 (211967_at), score: -0.57 TMEM2transmembrane protein 2 (218113_at), score: -0.63 TMEM30Atransmembrane protein 30A (217743_s_at), score: -0.65 TMEM39Btransmembrane protein 39B (218770_s_at), score: 0.66 TMF1TATA element modulatory factor 1 (213024_at), score: -0.67 TNIP1TNFAIP3 interacting protein 1 (207196_s_at), score: 0.63 TOMM20translocase of outer mitochondrial membrane 20 homolog (yeast) (200662_s_at), score: -0.63 TRIM21tripartite motif-containing 21 (204804_at), score: 0.69 TRIM24tripartite motif-containing 24 (213301_x_at), score: -0.62 TRMT2ATRM2 tRNA methyltransferase 2 homolog A (S. cerevisiae) (91617_at), score: -0.64 TRPC4transient receptor potential cation channel, subfamily C, member 4 (220817_at), score: -0.61 TSC2tuberous sclerosis 2 (215735_s_at), score: 0.66 TSKUtsukushin (218245_at), score: 0.69 TSNAXtranslin-associated factor X (203983_at), score: -0.57 TTC13tetratricopeptide repeat domain 13 (219481_at), score: -0.59 TUBA4Btubulin, alpha 4b (pseudogene) (207490_at), score: -0.78 UBA2ubiquitin-like modifier activating enzyme 2 (201177_s_at), score: -0.65 UBAC1UBA domain containing 1 (202151_s_at), score: 0.59 UBE2V2ubiquitin-conjugating enzyme E2 variant 2 (209096_at), score: -0.59 UBE2Wubiquitin-conjugating enzyme E2W (putative) (218521_s_at), score: -0.6 UFM1ubiquitin-fold modifier 1 (218050_at), score: -0.74 UGDHUDP-glucose dehydrogenase (203343_at), score: -0.7 UHRF1BP1LUHRF1 binding protein 1-like (213118_at), score: -0.64 URB1URB1 ribosome biogenesis 1 homolog (S. cerevisiae) (212996_s_at), score: -0.65 USF2upstream transcription factor 2, c-fos interacting (202152_x_at), score: 0.59 USP12ubiquitin specific peptidase 12 (213327_s_at), score: -0.57 USP21ubiquitin specific peptidase 21 (218367_x_at), score: 0.61 USP46ubiquitin specific peptidase 46 (203869_at), score: -0.67 VAT1vesicle amine transport protein 1 homolog (T. californica) (208626_s_at), score: 0.62 VEGFBvascular endothelial growth factor B (203683_s_at), score: 0.78 VPS13Cvacuolar protein sorting 13 homolog C (S. cerevisiae) (218396_at), score: -0.69 VPS37Bvacuolar protein sorting 37 homolog B (S. cerevisiae) (221704_s_at), score: 0.68 VRK3vaccinia related kinase 3 (221998_s_at), score: 0.62 WBP2WW domain binding protein 2 (209117_at), score: 0.72 WDR4WD repeat domain 4 (221632_s_at), score: -0.73 WDR42AWD repeat domain 42A (202249_s_at), score: 0.62 WDR44WD repeat domain 44 (219297_at), score: -0.62 WDR6WD repeat domain 6 (217734_s_at), score: 0.67 WRNWerner syndrome (205667_at), score: -0.57 WWTR1WW domain containing transcription regulator 1 (202132_at), score: -0.58 XPCxeroderma pigmentosum, complementation group C (209375_at), score: 0.61 YEATS4YEATS domain containing 4 (218911_at), score: -0.56 YTHDC2YTH domain containing 2 (213077_at), score: -0.68 YTHDF3YTH domain family, member 3 (221749_at), score: -0.64 ZBTB10zinc finger and BTB domain containing 10 (219312_s_at), score: -0.69 ZC3H14zinc finger CCCH-type containing 14 (213064_at), score: -0.63 ZCCHC10zinc finger, CCHC domain containing 10 (221193_s_at), score: -0.78 ZDHHC4zinc finger, DHHC-type containing 4 (220261_s_at), score: 0.58 ZDHHC7zinc finger, DHHC-type containing 7 (218606_at), score: 0.73 ZFAND1zinc finger, AN1-type domain 1 (218919_at), score: -0.58 ZFHX3zinc finger homeobox 3 (208033_s_at), score: 0.58 ZFYVE16zinc finger, FYVE domain containing 16 (203651_at), score: -0.69 ZNF107zinc finger protein 107 (205739_x_at), score: -0.65 ZNF23zinc finger protein 23 (KOX 16) (213934_s_at), score: -0.6 ZNF267zinc finger protein 267 (219540_at), score: -0.57 ZNF395zinc finger protein 395 (218149_s_at), score: 0.61 ZNF580zinc finger protein 580 (220748_s_at), score: 0.61 ZNF706zinc finger protein 706 (218059_at), score: 0.64
| Id | sample | Experiment | ExpName | Array | Syndrome | Cell.line |
|---|---|---|---|---|---|---|
| E-TABM-263-raw-cel-1515485891.cel | 13 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486371.cel | 37 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485811.cel | 9 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485771.cel | 7 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485751.cel | 6 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486091.cel | 23 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10358_WBS.CEL | 1 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515486291.cel | 33 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| 10590_WBS.CEL | 2 | 8 | WBS | hgu133plus2 | WBS | WBS 1 |
| E-TABM-263-raw-cel-1515485831.cel | 10 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485871.cel | 12 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515485691.cel | 3 | 6 | Cycle | hgu133a2 | none | Cycle 1 |
| E-TABM-263-raw-cel-1515486211.cel | 29 | 6 | Cycle | hgu133a2 | none | Cycle 1 |