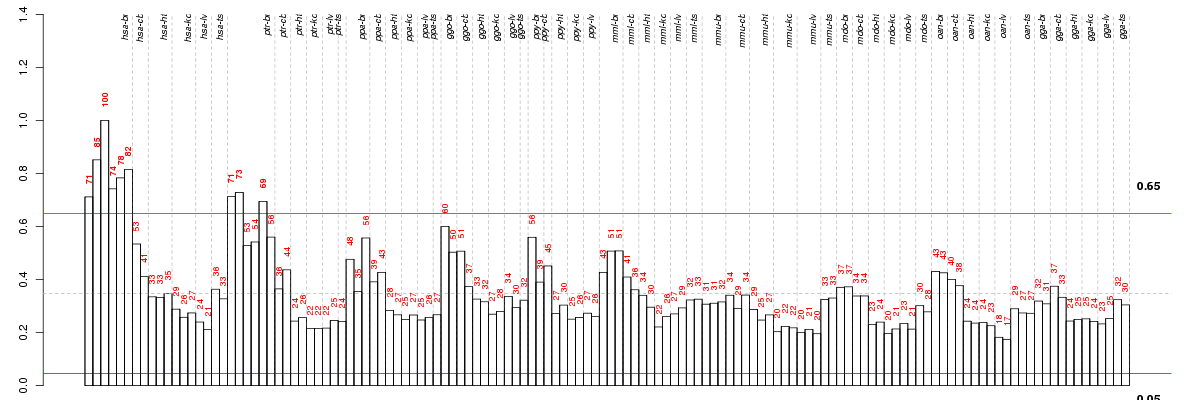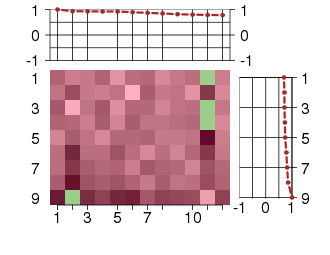

Under-expression is coded with green,
over-expression with red color.



C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.8 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.93 CAPN9calpain 9 (ENSG00000135773), score: 0.89 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 0.92 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.93 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.82 GLDNgliomedin (ENSG00000186417), score: 0.92 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 1 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.78 PMP2peripheral myelin protein 2 (ENSG00000147588), score: 0.87 RS1retinoschisin 1 (ENSG00000102104), score: 0.79 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.84
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_br_m5_ca1 | ptr | br | m | 5 |
| hsa_br_m2_ca1 | hsa | br | m | 2 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| hsa_br_m1_ca1 | hsa | br | m | 1 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |