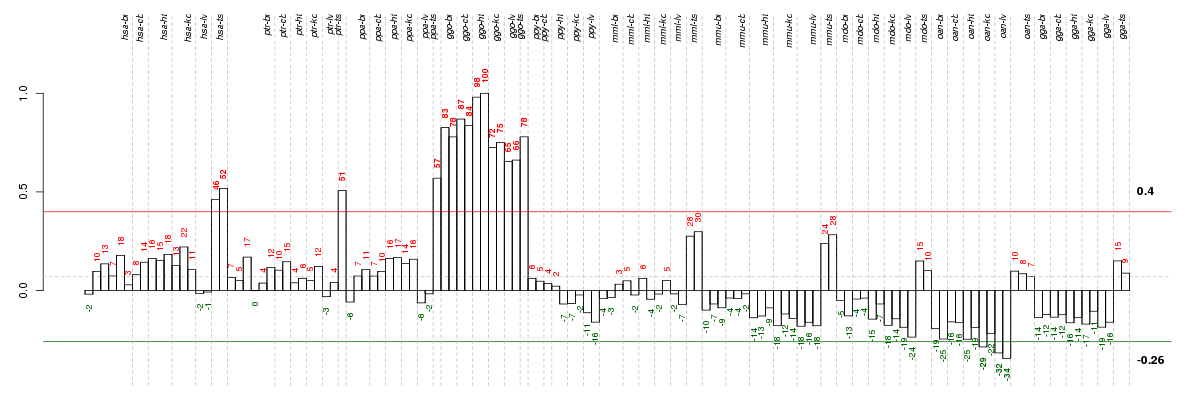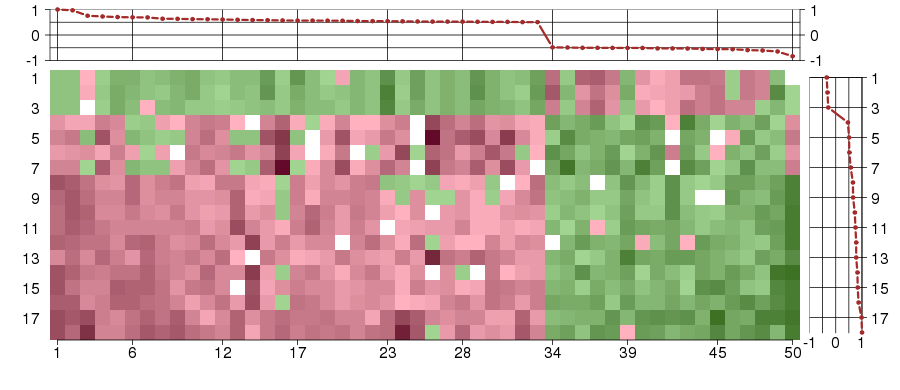

Under-expression is coded with green,
over-expression with red color.



ABCC1ATP-binding cassette, sub-family C (CFTR/MRP), member 1 (ENSG00000103222), score: 0.62 ANKRD54ankyrin repeat domain 54 (ENSG00000100124), score: -0.53 ARCN1archain 1 (ENSG00000095139), score: -0.59 BARX1BARX homeobox 1 (ENSG00000131668), score: 0.57 BEST1bestrophin 1 (ENSG00000167995), score: 0.55 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (ENSG00000156970), score: 0.62 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 1 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.97 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.5 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.56 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.64 CUL1cullin 1 (ENSG00000055130), score: -0.83 DCTdopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (ENSG00000080166), score: 0.54 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.6 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: -0.61 FAM59Bfamily with sequence similarity 59, member B (ENSG00000157833), score: 0.63 HAUS8HAUS augmin-like complex, subunit 8 (ENSG00000131351), score: 0.56 HESX1HESX homeobox 1 (ENSG00000163666), score: 0.52 HKDC1hexokinase domain containing 1 (ENSG00000156510), score: 0.69 IL17Binterleukin 17B (ENSG00000127743), score: 0.54 INHAinhibin, alpha (ENSG00000123999), score: 0.51 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.69 KCTD20potassium channel tetramerisation domain containing 20 (ENSG00000112078), score: -0.49 KIF15kinesin family member 15 (ENSG00000163808), score: 0.73 LHX4LIM homeobox 4 (ENSG00000121454), score: 0.51 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: 0.54 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: -0.48 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: 0.57 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.59 MYCBPAPMYCBP associated protein (ENSG00000136449), score: 0.53 NDFIP1Nedd4 family interacting protein 1 (ENSG00000131507), score: -0.51 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: -0.51 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: -0.56 PCID2PCI domain containing 2 (ENSG00000126226), score: 0.61 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.53 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.57 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.55 PRTGprotogenin (ENSG00000166450), score: 0.53 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: -0.55 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.76 PUS7pseudouridylate synthase 7 homolog (S. cerevisiae) (ENSG00000091127), score: -0.51 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: -0.65 RECQL4RecQ protein-like 4 (ENSG00000160957), score: 0.7 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: -0.5 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: 0.51 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: -0.53 TMEM41Btransmembrane protein 41B (ENSG00000166471), score: -0.5 UIMC1ubiquitin interaction motif containing 1 (ENSG00000087206), score: 0.52 VTA1Vps20-associated 1 homolog (S. cerevisiae) (ENSG00000009844), score: -0.53 WDR60WD repeat domain 60 (ENSG00000126870), score: 0.58
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| hsa_ts_m2_ca1 | hsa | ts | m | 2 |
| ptr_ts_m_ca1 | ptr | ts | m | _ |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| ppa_ts_m_ca1 | ppa | ts | m | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_ts_m_ca1 | ggo | ts | m | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |