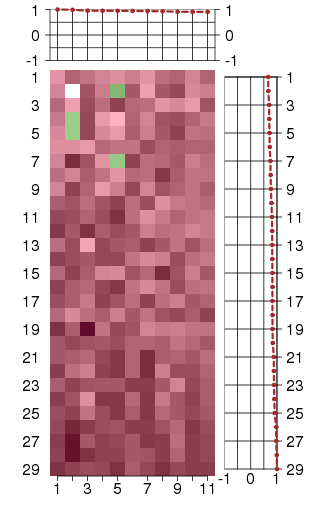

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 01100 | 2.047e-02 | 1.065 | 5 | 353 | Metabolic pathways |
| 00380 | 3.342e-02 | 0.04825 | 2 | 16 | Tryptophan metabolism |
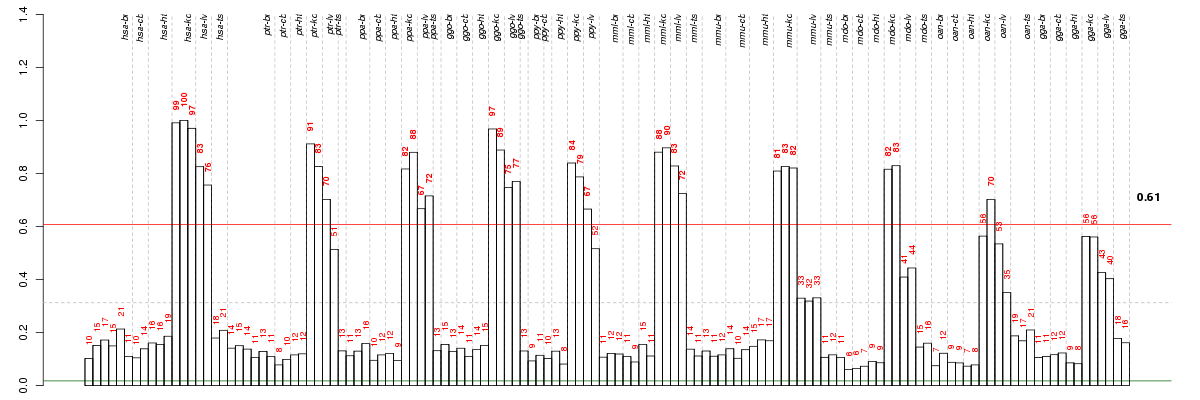
ACMSDaminocarboxymuconate semialdehyde decarboxylase (ENSG00000153086), score: 0.94 CRYL1crystallin, lambda 1 (ENSG00000165475), score: 0.91 DDCdopa decarboxylase (aromatic L-amino acid decarboxylase) (ENSG00000132437), score: 0.91 HAO2hydroxyacid oxidase 2 (long chain) (ENSG00000116882), score: 0.91 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.94 RAB17RAB17, member RAS oncogene family (ENSG00000124839), score: 0.93 SLC5A9solute carrier family 5 (sodium/glucose cotransporter), member 9 (ENSG00000117834), score: 0.95 SLC7A9solute carrier family 7 (cationic amino acid transporter, y+ system), member 9 (ENSG00000021488), score: 1 TMPRSS2transmembrane protease, serine 2 (ENSG00000184012), score: 0.95 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.95 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.99
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppy_lv_m_ca1 | ppy | lv | m | _ |
| ppa_lv_m_ca1 | ppa | lv | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| ptr_lv_m_ca1 | ptr | lv | m | _ |
| ppa_lv_f_ca1 | ppa | lv | f | _ |
| mml_lv_f_ca1 | mml | lv | f | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| ppy_kd_f_ca1 | ppy | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| ppy_kd_m_ca1 | ppy | kd | m | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| mml_kd_f_ca1 | mml | kd | f | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |