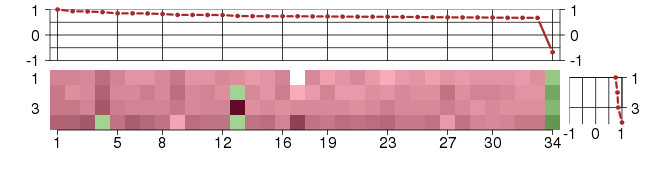

Under-expression is coded with green,
over-expression with red color.



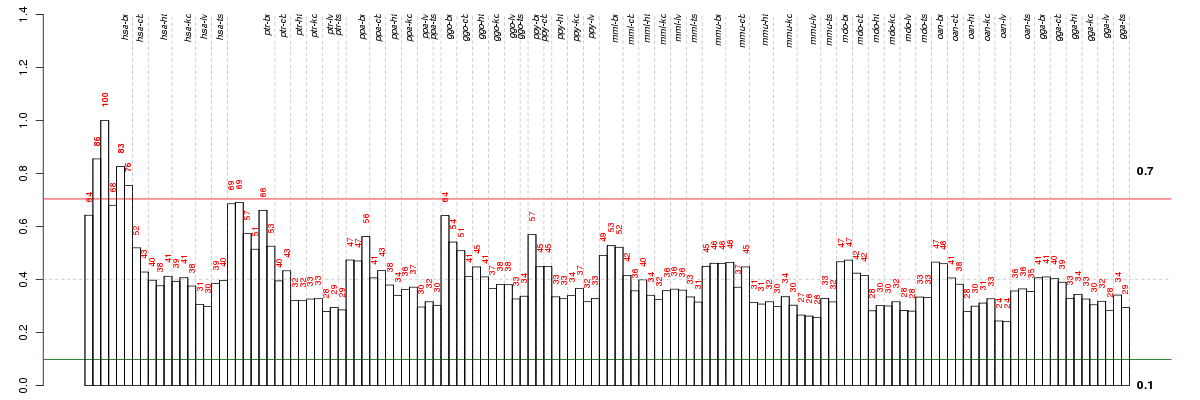
ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.71 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.68 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.71 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.9 CAPN9calpain 9 (ENSG00000135773), score: 0.85 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 1 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.93 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.71 DYNC1I2dynein, cytoplasmic 1, intermediate chain 2 (ENSG00000077380), score: 0.73 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.7 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.72 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.73 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.79 GLDNgliomedin (ENSG00000186417), score: 0.82 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.69 JAM3junctional adhesion molecule 3 (ENSG00000166086), score: 0.68 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.92 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.74 MSGN1mesogenin 1 (ENSG00000151379), score: 0.75 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.72 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.69 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.78 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.79 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.74 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.74 RAPGEF5Rap guanine nucleotide exchange factor (GEF) 5 (ENSG00000136237), score: 0.67 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.68 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.73 RS1retinoschisin 1 (ENSG00000102104), score: 0.79 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.69 SLC44A1solute carrier family 44, member 1 (ENSG00000070214), score: 0.72 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.68 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.84 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.85
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_f_ca1 | hsa | br | f | _ |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |