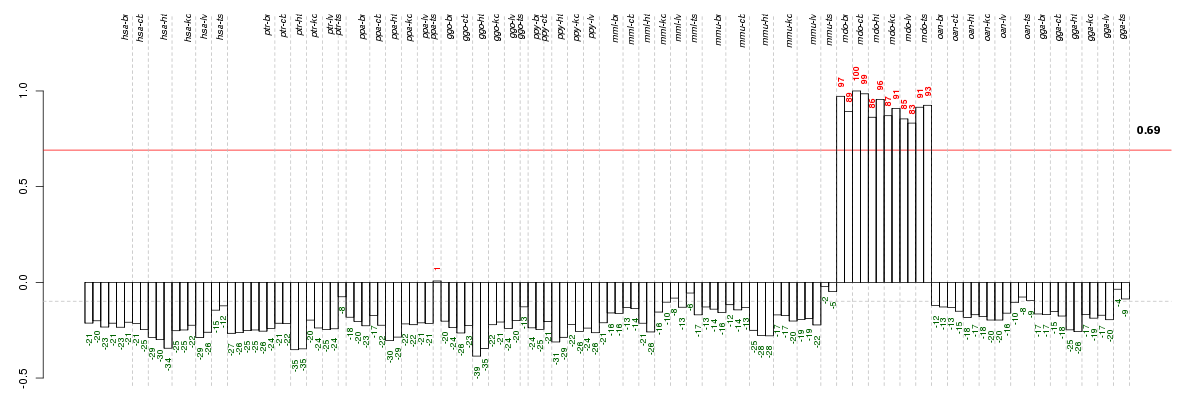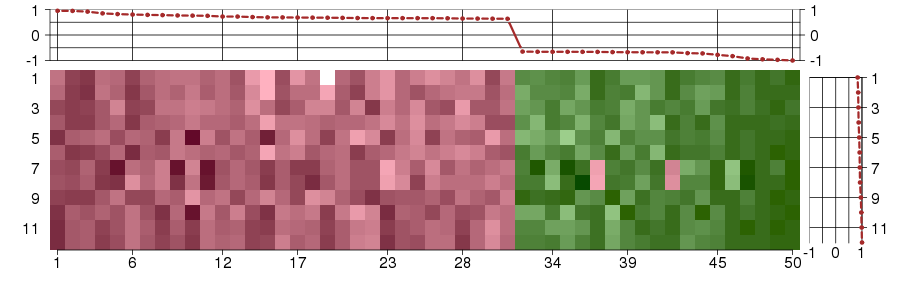

Under-expression is coded with green,
over-expression with red color.



ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.68 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.64 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.66 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.66 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.68 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.69 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.65 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.76 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.7 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.79 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.92 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.67 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.68 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.66 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.66 FBXO22F-box protein 22 (ENSG00000167196), score: 0.65 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.85 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.66 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.71 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.95 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.66 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.64 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.69 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.68 ND2MTND2 (ENSG00000198763), score: -0.95 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.92 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.72 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.64 PGRprogesterone receptor (ENSG00000082175), score: 0.68 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.72 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.66 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.75 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.78 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.67 SAP30LSAP30-like (ENSG00000164576), score: 0.66 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.66 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.94 SNX13sorting nexin 13 (ENSG00000071189), score: 0.66 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.66 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.78 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.68 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.8 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.77 TSNtranslin (ENSG00000211460), score: -0.66 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.83 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.68 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.97 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.82 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.72
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |