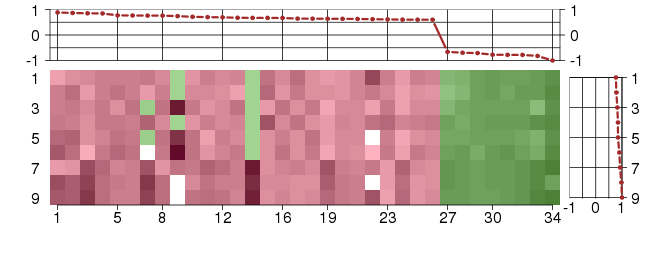

Under-expression is coded with green,
over-expression with red color.



ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.7 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.6 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.82 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.63 CCDC92coiled-coil domain containing 92 (ENSG00000119242), score: -0.69 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.84 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.86 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.85 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.62 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.61 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.67 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.63 H1FXH1 histone family, member X (ENSG00000184897), score: -1 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: -0.66 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.67 LIFRleukemia inhibitory factor receptor alpha (ENSG00000113594), score: 0.64 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.74 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.68 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.69 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.78 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.71 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.77 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.77 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.67 PRKD3protein kinase D3 (ENSG00000115825), score: 0.6 RNF139ring finger protein 139 (ENSG00000170881), score: 0.64 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.6 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.78 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.76 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.71 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.77 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.64 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.88 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.76
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |