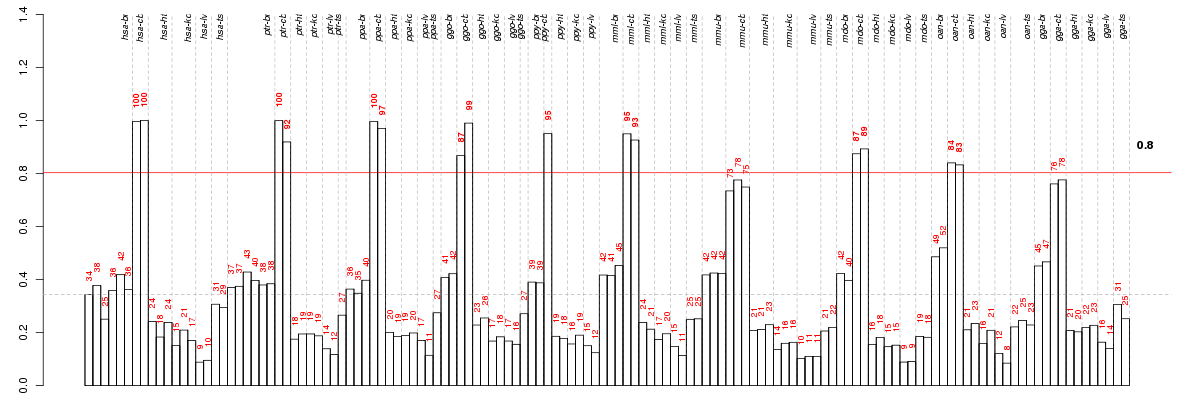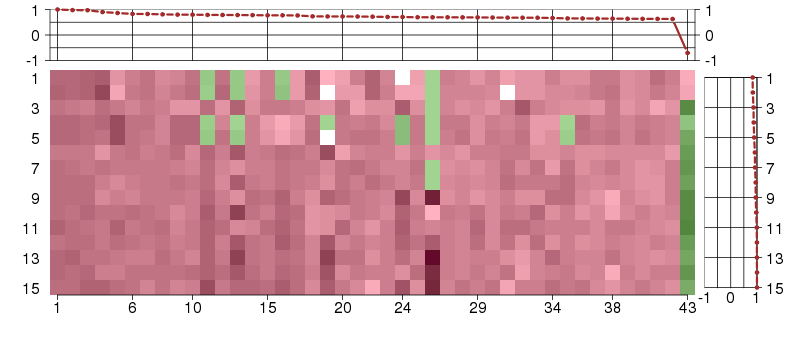

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
hedgehog receptor activity
Combining with the hedgehog protein to initiate a change in cell activity.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.63 C15orf27chromosome 15 open reading frame 27 (ENSG00000169758), score: 0.69 C6orf115chromosome 6 open reading frame 115 (ENSG00000146386), score: -0.7 C8orf79chromosome 8 open reading frame 79 (ENSG00000170941), score: 0.73 CBLN1cerebellin 1 precursor (ENSG00000102924), score: 0.82 CDH7cadherin 7, type 2 (ENSG00000081138), score: 0.7 CHRNA3cholinergic receptor, nicotinic, alpha 3 (ENSG00000080644), score: 0.73 CRTAMcytotoxic and regulatory T cell molecule (ENSG00000109943), score: 0.79 FAT2FAT tumor suppressor homolog 2 (Drosophila) (ENSG00000086570), score: 0.98 FGF5fibroblast growth factor 5 (ENSG00000138675), score: 0.86 FZD7frizzled homolog 7 (Drosophila) (ENSG00000155760), score: 0.69 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.73 GRM1glutamate receptor, metabotropic 1 (ENSG00000152822), score: 0.64 KCNC1potassium voltage-gated channel, Shaw-related subfamily, member 1 (ENSG00000129159), score: 0.64 KCNK10potassium channel, subfamily K, member 10 (ENSG00000100433), score: 0.68 KCNK9potassium channel, subfamily K, member 9 (ENSG00000169427), score: 0.77 LRCH1leucine-rich repeats and calponin homology (CH) domain containing 1 (ENSG00000136141), score: 0.68 LRRC38leucine rich repeat containing 38 (ENSG00000162494), score: 0.64 MAB21L1mab-21-like 1 (C. elegans) (ENSG00000180660), score: 0.97 MAML3mastermind-like 3 (Drosophila) (ENSG00000196782), score: 0.8 MDGA1MAM domain containing glycosylphosphatidylinositol anchor 1 (ENSG00000112139), score: 0.77 MEIS1Meis homeobox 1 (ENSG00000143995), score: 0.63 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.78 NKX6-3NK6 homeobox 3 (ENSG00000165066), score: 0.78 NRG2neuregulin 2 (ENSG00000158458), score: 0.65 PAX3paired box 3 (ENSG00000135903), score: 0.68 PAX6paired box 6 (ENSG00000007372), score: 0.83 PLCB4phospholipase C, beta 4 (ENSG00000101333), score: 0.63 PTCH1patched 1 (ENSG00000185920), score: 0.65 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.71 SKIv-ski sarcoma viral oncogene homolog (avian) (ENSG00000157933), score: 0.67 SKOR1SKI family transcriptional corepressor 1 (ENSG00000188779), score: 0.8 SLC35F4solute carrier family 35, member F4 (ENSG00000151812), score: 0.9 SPINK6serine peptidase inhibitor, Kazal type 6 (ENSG00000178172), score: 0.69 SPTBN5spectrin, beta, non-erythrocytic 5 (ENSG00000137877), score: 0.71 TIAM1T-cell lymphoma invasion and metastasis 1 (ENSG00000156299), score: 0.77 TLL1tolloid-like 1 (ENSG00000038295), score: 0.72 TRIM67tripartite motif-containing 67 (ENSG00000119283), score: 0.79 UPF0639UPF0639 protein (ENSG00000175985), score: 0.72 ZFPM2zinc finger protein, multitype 2 (ENSG00000169946), score: 0.68 ZIC4Zic family member 4 (ENSG00000174963), score: 1 ZNF238zinc finger protein 238 (ENSG00000179456), score: 0.69 ZNF521zinc finger protein 521 (ENSG00000198795), score: 0.81
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| mml_cb_f_ca1 | mml | cb | f | _ |
| mml_cb_m_ca1 | mml | cb | m | _ |
| ppy_cb_f_ca1 | ppy | cb | f | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| hsa_cb_m_ca1 | hsa | cb | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |
| hsa_cb_f_ca1 | hsa | cb | f | _ |