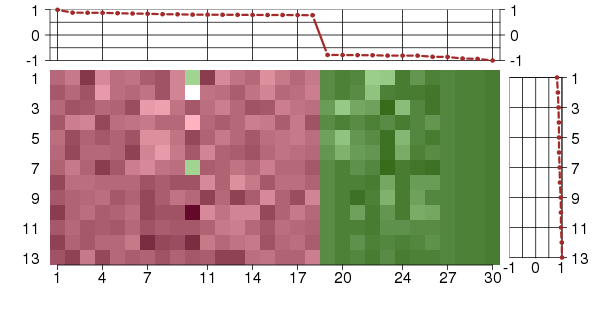

Under-expression is coded with green,
over-expression with red color.



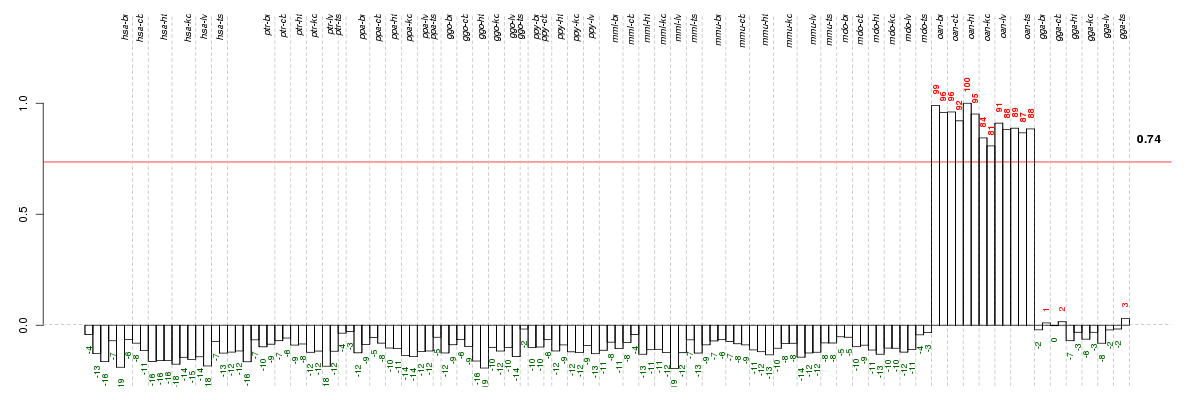
AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.83 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.78 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.81 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.82 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.79 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.81 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.81 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -1 CPLX4complexin 4 (ENSG00000166569), score: 0.8 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.78 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.79 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.88 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.81 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.8 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.84 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.79 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.86 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.78 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.87 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.87 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.79 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.85 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.94 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.78 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.92 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.99 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.85 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.78 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.79 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.78
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_ts_m1_ca1 | oan | ts | m | 1 |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_ts_m2_ca1 | oan | ts | m | 2 |
| oan_ts_m3_ca1 | oan | ts | m | 3 |
| oan_lv_m_ca1 | oan | lv | m | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_br_m_ca1 | oan | br | m | _ |
| oan_ht_m_ca1 | oan | ht | m | _ |