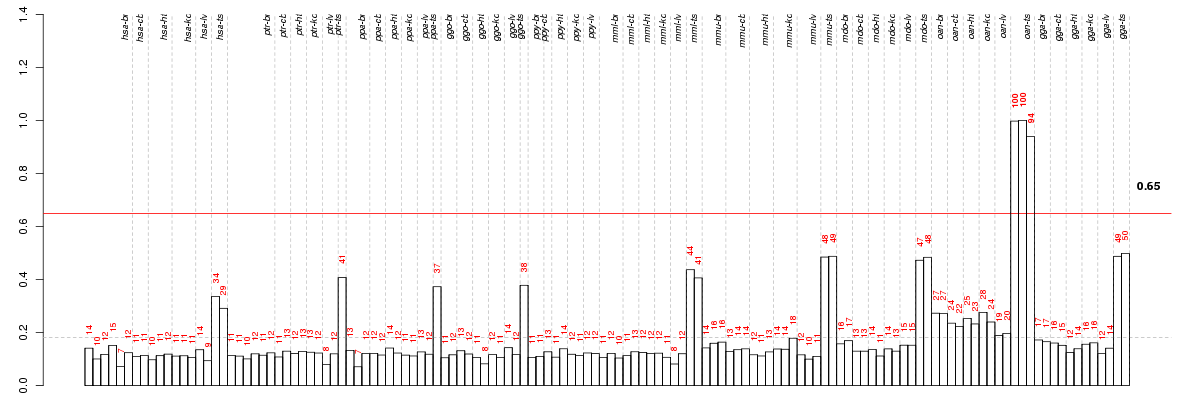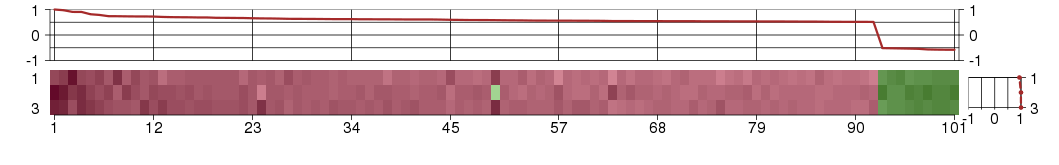

Under-expression is coded with green,
over-expression with red color.

M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.
mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
nuclear division
A process by which a cell nucleus is divided into two nuclei, with DNA and other nuclear contents distributed between the daughter nuclei.
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
cellular component organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cellular component.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
organelle fission
The creation of two or more organelles by division of one organelle.
all
NA
organelle organization
A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of an organelle within a cell. An organelle is an organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
mitosis
A cell cycle process comprising the steps by which the nucleus of a eukaryotic cell divides; the process involves condensation of chromosomal DNA into a highly compacted form. Canonically, mitosis produces two daughter nuclei whose chromosome complement is identical to that of the mother cell.
M phase of mitotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the mitotic cell cycle during which mitosis takes place.


ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (ENSG00000111875), score: 0.61 ASPMasp (abnormal spindle) homolog, microcephaly associated (Drosophila) (ENSG00000066279), score: 0.61 ATAD2ATPase family, AAA domain containing 2 (ENSG00000156802), score: 0.58 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.53 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.56 C10orf96chromosome 10 open reading frame 96 (ENSG00000182645), score: 0.52 C12orf48chromosome 12 open reading frame 48 (ENSG00000185480), score: 0.63 C13orf34chromosome 13 open reading frame 34 (ENSG00000136122), score: 0.53 C14orf39chromosome 14 open reading frame 39 (ENSG00000179008), score: 0.65 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.53 C1orf174chromosome 1 open reading frame 174 (ENSG00000198912), score: 0.73 C1orf9chromosome 1 open reading frame 9 (ENSG00000094975), score: 0.59 C5orf51chromosome 5 open reading frame 51 (ENSG00000205765), score: 0.69 C7orf60chromosome 7 open reading frame 60 (ENSG00000164603), score: 0.55 CAPN13calpain 13 (ENSG00000162949), score: 0.56 CASP2caspase 2, apoptosis-related cysteine peptidase (ENSG00000106144), score: 0.62 CENPIcentromere protein I (ENSG00000102384), score: 0.63 CHAF1Bchromatin assembly factor 1, subunit B (p60) (ENSG00000159259), score: 0.59 CHERPcalcium homeostasis endoplasmic reticulum protein (ENSG00000085872), score: -0.51 CNGA2cyclic nucleotide gated channel alpha 2 (ENSG00000183862), score: 0.9 COL17A1collagen, type XVII, alpha 1 (ENSG00000065618), score: 0.63 CPA1carboxypeptidase A1 (pancreatic) (ENSG00000091704), score: 0.67 CPA6carboxypeptidase A6 (ENSG00000165078), score: 0.73 CPLX4complexin 4 (ENSG00000166569), score: 0.52 CXorf30chromosome X open reading frame 30 (ENSG00000205081), score: 0.53 CYP11A1cytochrome P450, family 11, subfamily A, polypeptide 1 (ENSG00000140459), score: 0.63 DCLRE1CDNA cross-link repair 1C (ENSG00000152457), score: 0.61 DMRT3doublesex and mab-3 related transcription factor 3 (ENSG00000064218), score: 0.61 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.55 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.57 EFCAB1EF-hand calcium binding domain 1 (ENSG00000034239), score: 0.59 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (ENSG00000186871), score: 0.66 EXO1exonuclease 1 (ENSG00000174371), score: 0.52 EXOSC1exosome component 1 (ENSG00000171311), score: 0.55 EXOSC10exosome component 10 (ENSG00000171824), score: 0.74 FANCBFanconi anemia, complementation group B (ENSG00000181544), score: 1 FBXO21F-box protein 21 (ENSG00000135108), score: -0.52 FBXO47F-box protein 47 (ENSG00000204952), score: 0.56 GCM1glial cells missing homolog 1 (Drosophila) (ENSG00000137270), score: 0.56 GEMC1geminin coiled-coil domain-containing protein 1 (ENSG00000205835), score: 0.51 GPATCH2G patch domain containing 2 (ENSG00000092978), score: 0.53 GPR20G protein-coupled receptor 20 (ENSG00000204882), score: 0.79 GRXCR1glutaredoxin, cysteine rich 1 (ENSG00000215203), score: 0.72 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.56 HORMAD1HORMA domain containing 1 (ENSG00000143452), score: 0.52 HORMAD2HORMA domain containing 2 (ENSG00000176635), score: 0.54 ING3inhibitor of growth family, member 3 (ENSG00000071243), score: 0.53 KIAA0892KIAA0892 (ENSG00000129933), score: -0.57 KIF14kinesin family member 14 (ENSG00000118193), score: 0.54 LCORLligand dependent nuclear receptor corepressor-like (ENSG00000178177), score: 0.67 LCTlactase (ENSG00000115850), score: 0.74 LHCGRluteinizing hormone/choriogonadotropin receptor (ENSG00000138039), score: 0.73 LIN28Blin-28 homolog B (C. elegans) (ENSG00000187772), score: 0.61 LRGUKleucine-rich repeats and guanylate kinase domain containing (ENSG00000155530), score: 0.53 LRRC18leucine rich repeat containing 18 (ENSG00000165383), score: 0.56 LRRC52leucine rich repeat containing 52 (ENSG00000162763), score: 0.62 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.55 LYPD2LY6/PLAUR domain containing 2 (ENSG00000197353), score: 0.53 MELKmaternal embryonic leucine zipper kinase (ENSG00000165304), score: 0.54 MTF2metal response element binding transcription factor 2 (ENSG00000143033), score: 0.69 MYO18Amyosin XVIIIA (ENSG00000196535), score: -0.58 NBR1neighbor of BRCA1 gene 1 (ENSG00000188554), score: 0.53 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.54 NPRL2nitrogen permease regulator-like 2 (S. cerevisiae) (ENSG00000114388), score: -0.58 NR5A1nuclear receptor subfamily 5, group A, member 1 (ENSG00000136931), score: 0.57 ORC3Lorigin recognition complex, subunit 3-like (yeast) (ENSG00000135336), score: 0.7 OXTRoxytocin receptor (ENSG00000180914), score: 0.69 PAX5paired box 5 (ENSG00000196092), score: 0.67 PAX9paired box 9 (ENSG00000198807), score: 0.71 PHEXphosphate regulating endopeptidase homolog, X-linked (ENSG00000102174), score: 0.55 PI15peptidase inhibitor 15 (ENSG00000137558), score: 0.6 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.6 PPYR1pancreatic polypeptide receptor 1 (ENSG00000204174), score: 0.65 PSMD6proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 (ENSG00000163636), score: -0.53 RAD54BRAD54 homolog B (S. cerevisiae) (ENSG00000197275), score: 0.67 RASEFRAS and EF-hand domain containing (ENSG00000165105), score: 0.54 RBM46RNA binding motif protein 46 (ENSG00000151962), score: 0.56 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.91 RIBC2RIB43A domain with coiled-coils 2 (ENSG00000128408), score: 0.51 RPAP2RNA polymerase II associated protein 2 (ENSG00000122484), score: 0.62 SGOL1shugoshin-like 1 (S. pombe) (ENSG00000129810), score: 0.62 SLC6A14solute carrier family 6 (amino acid transporter), member 14 (ENSG00000087916), score: 0.97 SPC25SPC25, NDC80 kinetochore complex component, homolog (S. cerevisiae) (ENSG00000152253), score: 0.54 STRA8stimulated by retinoic acid gene 8 homolog (mouse) (ENSG00000146857), score: 0.63 TAAR2trace amine associated receptor 2 (ENSG00000146378), score: 0.58 TBX4T-box 4 (ENSG00000121075), score: 0.81 TDRD1tudor domain containing 1 (ENSG00000095627), score: 0.53 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.54 TRPC6transient receptor potential cation channel, subfamily C, member 6 (ENSG00000137672), score: 0.62 TTC27tetratricopeptide repeat domain 27 (ENSG00000018699), score: 0.62 TTKTTK protein kinase (ENSG00000112742), score: 0.54 UBE4Aubiquitination factor E4A (UFD2 homolog, yeast) (ENSG00000110344), score: 0.64 USP49ubiquitin specific peptidase 49 (ENSG00000164663), score: 0.54 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.52 WDR78WD repeat domain 78 (ENSG00000152763), score: 0.57 ZBTB40zinc finger and BTB domain containing 40 (ENSG00000184677), score: 0.54 ZNF366zinc finger protein 366 (ENSG00000178175), score: 0.61 ZNF438zinc finger protein 438 (ENSG00000183621), score: 0.7 ZNF644zinc finger protein 644 (ENSG00000122482), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_ts_m2_ca1 | oan | ts | m | 2 |
| oan_ts_m3_ca1 | oan | ts | m | 3 |
| oan_ts_m1_ca1 | oan | ts | m | 1 |