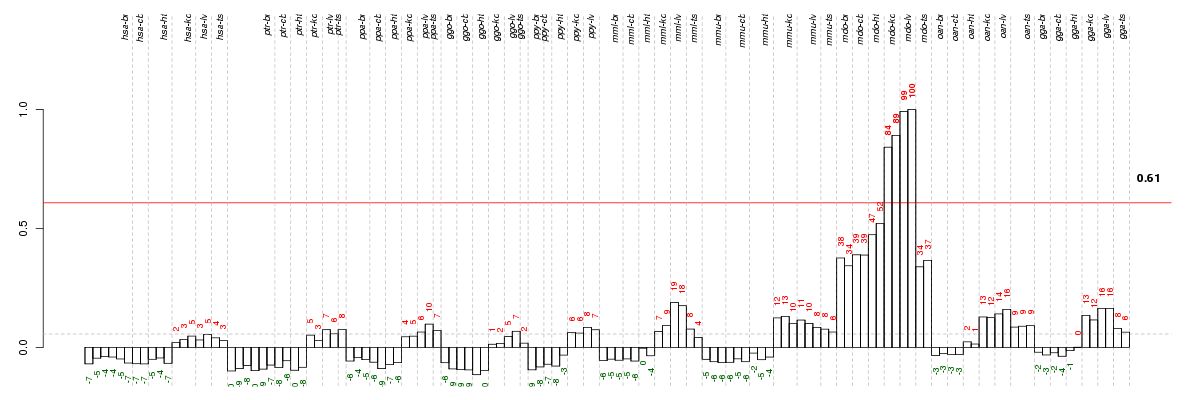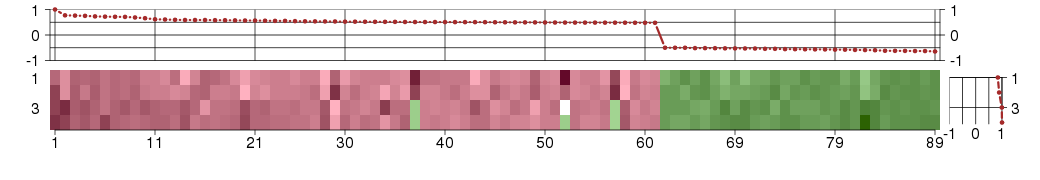

Under-expression is coded with green,
over-expression with red color.



AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.5 ABHD6abhydrolase domain containing 6 (ENSG00000163686), score: -0.57 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.52 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.49 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.62 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.54 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.52 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.56 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.5 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 1 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.57 ATMINATM interactor (ENSG00000166454), score: -0.58 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.51 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.55 ATXN2ataxin 2 (ENSG00000204842), score: 0.5 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.48 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.53 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.57 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.52 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.5 CLTBclathrin, light chain B (ENSG00000175416), score: -0.63 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.56 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.52 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.52 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.55 CYTBcytochrome b (ENSG00000198727), score: -0.65 CYTH1cytohesin 1 (ENSG00000108669), score: -0.56 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.5 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.6 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.56 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: 0.51 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.49 EPYCepiphycan (ENSG00000083782), score: 0.53 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.75 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.66 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.51 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.48 GABRPgamma-aminobutyric acid (GABA) A receptor, pi (ENSG00000094755), score: 0.49 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.61 GSCgoosecoid homeobox (ENSG00000133937), score: 0.48 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.53 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.51 IMPAD1inositol monophosphatase domain containing 1 (ENSG00000104331), score: -0.59 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.5 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: 0.53 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.58 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.71 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.53 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.49 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.54 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.49 ND2MTND2 (ENSG00000198763), score: -0.62 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.5 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.71 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.51 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.59 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.52 PDP2pyruvate dehyrogenase phosphatase catalytic subunit 2 (ENSG00000172840), score: 0.59 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.54 PODNpodocan (ENSG00000174348), score: 0.62 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.77 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.48 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.52 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.5 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.48 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.49 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.52 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: 0.51 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.69 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.57 SNX2sorting nexin 2 (ENSG00000205302), score: 0.58 SPATS2spermatogenesis associated, serine-rich 2 (ENSG00000123352), score: -0.5 STAB2stabilin 2 (ENSG00000136011), score: 0.52 STAMBPSTAM binding protein (ENSG00000124356), score: 0.5 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.59 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.57 TEX264testis expressed 264 (ENSG00000164081), score: 0.49 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.73 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.48 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.55 TTF2transcription termination factor, RNA polymerase II (ENSG00000116830), score: 0.76 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.58 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.59 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.63 WDR37WD repeat domain 37 (ENSG00000047056), score: -0.53 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.62 ZDHHC13zinc finger, DHHC-type containing 13 (ENSG00000177054), score: 0.72 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.54 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.51
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |