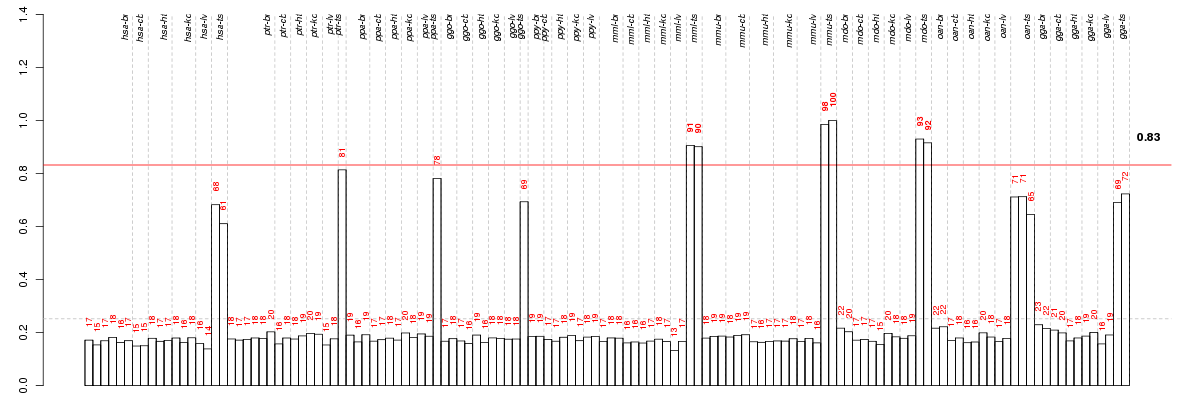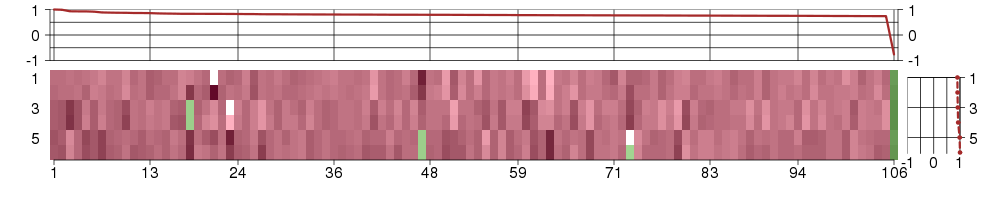

Under-expression is coded with green,
over-expression with red color.

reproduction
The production by an organism of new individuals that contain some portion of their genetic material inherited from that organism.
M phase
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the cell cycle comprising nuclear division.
microtubule-based process
Any cellular process that depends upon or alters the microtubule cytoskeleton, that part of the cytoskeleton comprising microtubules and their associated proteins.
microtubule-based movement
Movement of organelles, other microtubules and other particles along microtubules, mediated by motor proteins.
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
meiosis I
A cell cycle process comprising the steps by which a cell progresses through the first phase of meiosis, in which cells divide and homologous chromosomes are paired and segregated from each other, producing two daughter cells.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
spermatogenesis
The process of formation of spermatozoa, including spermatocytogenesis and spermiogenesis.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
sexual reproduction
The regular alternation, in the life cycle of haplontic, diplontic and diplohaplontic organisms, of meiosis and fertilization which provides for the production offspring. In diplontic organisms there is a life cycle in which the products of meiosis behave directly as gametes, fusing to form a zygote from which the diploid, or sexually reproductive polyploid, adult organism will develop. In diplohaplontic organisms a haploid phase (gametophyte) exists in the life cycle between meiosis and fertilization (e.g. higher plants, many algae and Fungi); the products of meiosis are spores that develop as haploid individuals from which haploid gametes develop to form a diploid zygote; diplohaplontic organisms show an alternation of haploid and diploid generations. In haplontic organisms meiosis occurs in the zygote, giving rise to four haploid cells (e.g. many algae and protozoa), only the zygote is diploid and this may form a resistant spore, tiding organisms over hard times.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
cell cycle phase
A cell cycle process comprising the steps by which a cell progresses through one of the biochemical and morphological phases and events that occur during successive cell replication or nuclear replication events.
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
male gamete generation
Generation of the male gamete; specialised haploid cells produced by meiosis and along with a female gamete takes part in sexual reproduction.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
cell division
The process resulting in the physical partitioning and separation of a cell into daughter cells.
meiotic cell cycle
Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
all
NA
reproductive process
A biological process that directly contributes to the process of producing new individuals by one or two organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
multicellular organism reproduction
The biological process by which new individuals are produced by one or two multicellular organisms. The new individuals inherit some proportion of their genetic material from the parent or parents.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
reproductive process in a multicellular organism
The process, occurring above the cellular level, that is pertinent to the reproductive function of a multicellular organism. This includes the integrated processes at the level of tissues and organs.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
gamete generation
The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell.
meiosis
A cell cycle process comprising the steps by which a cell progresses through the nuclear division phase of a meiotic cell cycle, the specialized nuclear and cell division in which a single diploid cell undergoes two nuclear divisions following a single round of DNA replication in order to produce four daughter cells that contain half the number of chromosomes as the diploid cell. Meiotic division occurs during the formation of gametes from diploid organisms and at the beginning of haplophase in those organisms that alternate between diploid and haploid generations.
M phase of meiotic cell cycle
A cell cycle process comprising the steps by which a cell progresses through M phase, the part of the meiotic cell cycle during which meiosis takes place.
meiosis I
A cell cycle process comprising the steps by which a cell progresses through the first phase of meiosis, in which cells divide and homologous chromosomes are paired and segregated from each other, producing two daughter cells.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
microtubule organizing center
A cytoplasmic structure that can catalyze gamma-tubulin-dependent microtubule nucleation and that can anchor microtubules by interacting with their minus ends, plus ends or sides.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
microtubule cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of microtubules and associated proteins.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
pole plasm
Differentiated cytoplasm associated with a pole (animal, vegetal, anterior, or posterior) of an oocyte, egg or early embryo.
germ plasm
Differentiated cytoplasm associated with a pole of an oocyte, egg or early embryo that will be inherited by the cells that will give rise to the germ line.
piP-body
A P granule that contains the PIWIL4-TDRD9 module, a set of proteins that act in the secondary piRNA pathway.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
microtubule organizing center
A cytoplasmic structure that can catalyze gamma-tubulin-dependent microtubule nucleation and that can anchor microtubules by interacting with their minus ends, plus ends or sides.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.
cytoskeletal part
Any constituent part of the cytoskeleton, a cellular scaffolding or skeleton that maintains cell shape, enables some cell motion (using structures such as flagella and cilia), and plays important roles in both intra-cellular transport (e.g. the movement of vesicles and organelles) and cellular division. Includes constituent parts of intermediate filaments, microfilaments, microtubules, and the microtrabecular lattice.
microtubule organizing center
A cytoplasmic structure that can catalyze gamma-tubulin-dependent microtubule nucleation and that can anchor microtubules by interacting with their minus ends, plus ends or sides.
spindle
The array of microtubules and associated molecules that forms between opposite poles of a eukaryotic cell during mitosis or meiosis and serves to move the duplicated chromosomes apart.
microtubule
Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle.
P granule
A small cytoplasmic, non-membranous RNA/protein complex aggregates in the primordial germ cells of many higher eukaryotes.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
motor activity
Catalysis of movement along a polymeric molecule such as a microfilament or microtubule, coupled to the hydrolysis of a nucleoside triphosphate.
microtubule motor activity
Catalysis of movement along a microtubule, coupled to the hydrolysis of a nucleoside triphosphate (usually ATP).
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
nucleoside-triphosphatase activity
Catalysis of the reaction: a nucleoside triphosphate + H2O = nucleoside diphosphate + phosphate.
pyrophosphatase activity
Catalysis of the hydrolysis of a pyrophosphate bond between two phosphate groups, leaving one phosphate on each of the two fragments.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on acid anhydrides
Catalysis of the hydrolysis of any acid anhydride.
hydrolase activity, acting on acid anhydrides, in phosphorus-containing anhydrides
Catalysis of the hydrolysis of any acid anhydride which contains phosphorus.
all
NA
ACVR1activin A receptor, type I (ENSG00000115170), score: -0.76 ADAD1adenosine deaminase domain containing 1 (testis-specific) (ENSG00000164113), score: 0.77 AGBL2ATP/GTP binding protein-like 2 (ENSG00000165923), score: 0.76 AGBL3ATP/GTP binding protein-like 3 (ENSG00000146856), score: 0.76 AK7adenylate kinase 7 (ENSG00000140057), score: 0.75 ARID2AT rich interactive domain 2 (ARID, RFX-like) (ENSG00000189079), score: 0.81 AURKAaurora kinase A (ENSG00000087586), score: 0.75 C11orf82chromosome 11 open reading frame 82 (ENSG00000165490), score: 0.93 C12orf50chromosome 12 open reading frame 50 (ENSG00000165805), score: 0.78 C12orf63chromosome 12 open reading frame 63 (ENSG00000188596), score: 0.83 C13orf34chromosome 13 open reading frame 34 (ENSG00000136122), score: 0.74 C14orf166Bchromosome 14 open reading frame 166B (ENSG00000100565), score: 0.88 C15orf26chromosome 15 open reading frame 26 (ENSG00000156206), score: 0.86 C17orf104chromosome 17 open reading frame 104 (ENSG00000180336), score: 0.76 C3orf38chromosome 3 open reading frame 38 (ENSG00000179021), score: 0.77 C6orf204chromosome 6 open reading frame 204 (ENSG00000111860), score: 0.8 C7orf31chromosome 7 open reading frame 31 (ENSG00000153790), score: 0.78 C7orf45chromosome 7 open reading frame 45 (ENSG00000165120), score: 0.86 C9orf96chromosome 9 open reading frame 96 (ENSG00000198870), score: 0.76 C9orf98chromosome 9 open reading frame 98 (ENSG00000165695), score: 0.79 CCDC108coiled-coil domain containing 108 (ENSG00000181378), score: 0.79 CCDC135coiled-coil domain containing 135 (ENSG00000159625), score: 0.76 CCDC27coiled-coil domain containing 27 (ENSG00000162592), score: 0.75 CCDC30coiled-coil domain containing 30 (ENSG00000186409), score: 0.74 CCDC37coiled-coil domain containing 37 (ENSG00000163885), score: 0.84 CCDC41coiled-coil domain containing 41 (ENSG00000173588), score: 0.79 CCDC63coiled-coil domain containing 63 (ENSG00000173093), score: 0.82 CCDC67coiled-coil domain containing 67 (ENSG00000165325), score: 0.82 CCDC83coiled-coil domain containing 83 (ENSG00000150676), score: 0.86 CCR4chemokine (C-C motif) receptor 4 (ENSG00000183813), score: 0.83 CDC14ACDC14 cell division cycle 14 homolog A (S. cerevisiae) (ENSG00000079335), score: 0.8 CENPFcentromere protein F, 350/400kDa (mitosin) (ENSG00000117724), score: 0.77 CEP152centrosomal protein 152kDa (ENSG00000103995), score: 0.79 CEP350centrosomal protein 350kDa (ENSG00000135837), score: 0.81 CEP55centrosomal protein 55kDa (ENSG00000138180), score: 0.77 CPEB2cytoplasmic polyadenylation element binding protein 2 (ENSG00000137449), score: 0.77 DDX20DEAD (Asp-Glu-Ala-Asp) box polypeptide 20 (ENSG00000064703), score: 0.77 DDX4DEAD (Asp-Glu-Ala-Asp) box polypeptide 4 (ENSG00000152670), score: 0.76 DIS3LDIS3 mitotic control homolog (S. cerevisiae)-like (ENSG00000166938), score: 0.76 DNAH8dynein, axonemal, heavy chain 8 (ENSG00000124721), score: 0.81 DNAJC5BDnaJ (Hsp40) homolog, subfamily C, member 5 beta (ENSG00000147570), score: 0.84 DTLdenticleless homolog (Drosophila) (ENSG00000143476), score: 0.74 DUSP13dual specificity phosphatase 13 (ENSG00000079393), score: 0.76 E2F8E2F transcription factor 8 (ENSG00000129173), score: 0.78 EFCAB5EF-hand calcium binding domain 5 (ENSG00000176927), score: 0.79 ENO4enolase family member 4 (ENSG00000188316), score: 0.77 FAM161Afamily with sequence similarity 161, member A (ENSG00000170264), score: 0.99 FAM194Afamily with sequence similarity 194, member A (ENSG00000163645), score: 0.77 FAM46Cfamily with sequence similarity 46, member C (ENSG00000183508), score: 0.74 FAM54Afamily with sequence similarity 54, member A (ENSG00000146410), score: 0.76 FAM81Bfamily with sequence similarity 81, member B (ENSG00000153347), score: 0.8 FBXO43F-box protein 43 (ENSG00000156509), score: 0.8 FEM1Cfem-1 homolog c (C. elegans) (ENSG00000145780), score: 0.75 GDPD4glycerophosphodiester phosphodiesterase domain containing 4 (ENSG00000178795), score: 0.93 GEMIN5gem (nuclear organelle) associated protein 5 (ENSG00000082516), score: 0.78 GKAP1G kinase anchoring protein 1 (ENSG00000165113), score: 0.8 IQCHIQ motif containing H (ENSG00000103599), score: 0.81 KIAA0586KIAA0586 (ENSG00000100578), score: 0.79 KIAA1009KIAA1009 (ENSG00000135315), score: 0.75 KIF14kinesin family member 14 (ENSG00000118193), score: 0.79 KIF18Bkinesin family member 18B (ENSG00000186185), score: 0.79 KIF24kinesin family member 24 (ENSG00000186638), score: 0.74 KIF27kinesin family member 27 (ENSG00000165115), score: 0.87 KIF2Bkinesin family member 2B (ENSG00000141200), score: 0.75 KLHL10kelch-like 10 (Drosophila) (ENSG00000161594), score: 0.75 KNTC1kinetochore associated 1 (ENSG00000184445), score: 0.83 LASS3LAG1 homolog, ceramide synthase 3 (ENSG00000154227), score: 0.79 LGSNlengsin, lens protein with glutamine synthetase domain (ENSG00000146166), score: 0.83 LIG3ligase III, DNA, ATP-dependent (ENSG00000005156), score: 0.78 LRRC18leucine rich repeat containing 18 (ENSG00000165383), score: 0.75 LRRC50leucine rich repeat containing 50 (ENSG00000154099), score: 0.74 LRRIQ1leucine-rich repeats and IQ motif containing 1 (ENSG00000133640), score: 0.77 LRRIQ4leucine-rich repeats and IQ motif containing 4 (ENSG00000188306), score: 0.81 MAELmaelstrom homolog (Drosophila) (ENSG00000143194), score: 0.75 MAKmale germ cell-associated kinase (ENSG00000111837), score: 0.8 NRD1nardilysin (N-arginine dibasic convertase) (ENSG00000078618), score: 0.79 NUP153nucleoporin 153kDa (ENSG00000124789), score: 1 PDZD8PDZ domain containing 8 (ENSG00000165650), score: 0.82 RNF32ring finger protein 32 (ENSG00000105982), score: 0.78 ROPN1Lropporin 1-like (ENSG00000145491), score: 0.77 SERPINB12serpin peptidase inhibitor, clade B (ovalbumin), member 12 (ENSG00000166634), score: 0.79 SHCBP1SHC SH2-domain binding protein 1 (ENSG00000171241), score: 0.8 SMC1Bstructural maintenance of chromosomes 1B (ENSG00000077935), score: 0.83 SPATA17spermatogenesis associated 17 (ENSG00000162814), score: 0.76 SPATA18spermatogenesis associated 18 homolog (rat) (ENSG00000163071), score: 0.8 SPO11SPO11 meiotic protein covalently bound to DSB homolog (S. cerevisiae) (ENSG00000054796), score: 0.78 STILSCL/TAL1 interrupting locus (ENSG00000123473), score: 0.78 SYCP1synaptonemal complex protein 1 (ENSG00000198765), score: 0.75 TDRD5tudor domain containing 5 (ENSG00000162782), score: 0.81 TEX2testis expressed 2 (ENSG00000136478), score: 0.77 TMC1transmembrane channel-like 1 (ENSG00000165091), score: 0.83 TMC5transmembrane channel-like 5 (ENSG00000103534), score: 0.77 TMF1TATA element modulatory factor 1 (ENSG00000144747), score: 0.76 TMPRSS7transmembrane protease, serine 7 (ENSG00000176040), score: 0.77 TRIP12thyroid hormone receptor interactor 12 (ENSG00000153827), score: 0.81 TTC16tetratricopeptide repeat domain 16 (ENSG00000167094), score: 0.85 TTC29tetratricopeptide repeat domain 29 (ENSG00000137473), score: 0.79 UHRF1ubiquitin-like with PHD and ring finger domains 1 (ENSG00000034063), score: 0.8 VWA5B1von Willebrand factor A domain containing 5B1 (ENSG00000158816), score: 0.93 WDR48WD repeat domain 48 (ENSG00000114742), score: 0.87 WDR64WD repeat domain 64 (ENSG00000162843), score: 0.92 ZFAND3zinc finger, AN1-type domain 3 (ENSG00000156639), score: 0.74 ZNRF4zinc and ring finger 4 (ENSG00000105428), score: 0.78 ZPBP2zona pellucida binding protein 2 (ENSG00000186075), score: 0.8
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mml_ts_m2_ca1 | mml | ts | m | 2 |
| mml_ts_m1_ca1 | mml | ts | m | 1 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mmu_ts_m1_ca1 | mmu | ts | m | 1 |
| mmu_ts_m2_ca1 | mmu | ts | m | 2 |