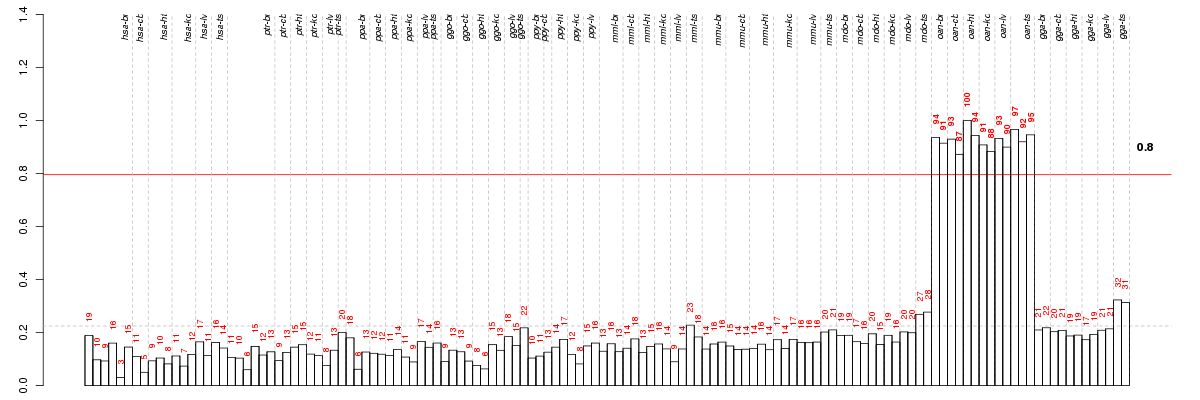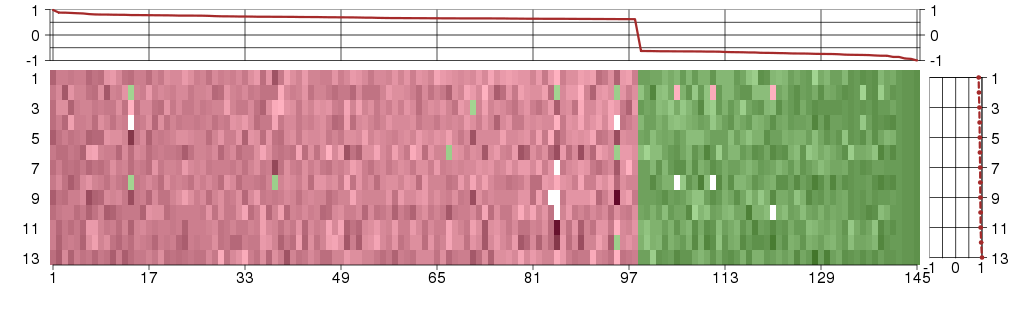

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.82 AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: 0.65 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.66 ACTN4actinin, alpha 4 (ENSG00000130402), score: -0.7 ADALadenosine deaminase-like (ENSG00000168803), score: 0.66 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.75 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.78 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: -0.68 ARHGEF18Rho/Rac guanine nucleotide exchange factor (GEF) 18 (ENSG00000104880), score: -0.67 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (ENSG00000163947), score: -0.73 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (ENSG00000111875), score: 0.76 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.76 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.69 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.73 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.7 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.64 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.8 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.71 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.81 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.79 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.81 CCDC93coiled-coil domain containing 93 (ENSG00000125633), score: 0.73 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.66 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.8 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.63 CHAF1Bchromatin assembly factor 1, subunit B (p60) (ENSG00000159259), score: 0.69 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.72 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.74 COG2component of oligomeric golgi complex 2 (ENSG00000135775), score: 0.65 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -1 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (ENSG00000173085), score: 0.67 CPLX4complexin 4 (ENSG00000166569), score: 0.78 CRKv-crk sarcoma virus CT10 oncogene homolog (avian) (ENSG00000167193), score: -0.64 CXorf23chromosome X open reading frame 23 (ENSG00000173681), score: 0.66 DAZAP1DAZ associated protein 1 (ENSG00000071626), score: -0.64 DCLRE1CDNA cross-link repair 1C (ENSG00000152457), score: 0.64 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.78 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.67 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.8 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.88 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.81 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.72 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (ENSG00000186871), score: 0.65 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.72 EXO1exonuclease 1 (ENSG00000174371), score: 0.64 EXOSC1exosome component 1 (ENSG00000171311), score: 0.7 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: -0.63 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.74 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: -0.63 FLNBfilamin B, beta (ENSG00000136068), score: -0.65 FOXP1forkhead box P1 (ENSG00000114861), score: -0.64 GOLGA1golgin A1 (ENSG00000136935), score: -0.65 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.8 GPN3GPN-loop GTPase 3 (ENSG00000111231), score: -0.68 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.68 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.65 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.85 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.63 HRH4histamine receptor H4 (ENSG00000134489), score: 0.71 HSPB11heat shock protein family B (small), member 11 (ENSG00000081870), score: 0.65 HTThuntingtin (ENSG00000197386), score: -0.73 INTS9integrator complex subunit 9 (ENSG00000104299), score: 0.65 KIAA0907KIAA0907 (ENSG00000132680), score: 0.65 KIAA1407KIAA1407 (ENSG00000163617), score: 0.76 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.65 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.71 LOH12CR1loss of heterozygosity, 12, chromosomal region 1 (ENSG00000165714), score: 0.77 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.79 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.64 MAP2K5mitogen-activated protein kinase kinase 5 (ENSG00000137764), score: -0.65 MCM6minichromosome maintenance complex component 6 (ENSG00000076003), score: 0.64 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.7 MLLT1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENSG00000130382), score: -0.71 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.86 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.62 MYNNmyoneurin (ENSG00000085274), score: 0.72 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.68 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: 0.73 NPRL2nitrogen permease regulator-like 2 (S. cerevisiae) (ENSG00000114388), score: -0.7 NUP133nucleoporin 133kDa (ENSG00000069248), score: 0.66 NUP37nucleoporin 37kDa (ENSG00000075188), score: 0.63 ORC3Lorigin recognition complex, subunit 3-like (yeast) (ENSG00000135336), score: 0.63 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.64 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.7 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.78 PLDNpallidin homolog (mouse) (ENSG00000104164), score: 0.66 POLGpolymerase (DNA directed), gamma (ENSG00000140521), score: -0.7 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.87 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.63 PUS1pseudouridylate synthase 1 (ENSG00000177192), score: -0.64 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.88 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.62 RANBP3RAN binding protein 3 (ENSG00000031823), score: -0.77 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.63 RCHY1ring finger and CHY zinc finger domain containing 1 (ENSG00000163743), score: 0.71 RECKreversion-inducing-cysteine-rich protein with kazal motifs (ENSG00000122707), score: 0.69 RFC3replication factor C (activator 1) 3, 38kDa (ENSG00000133119), score: 0.63 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: 0.63 RNF185ring finger protein 185 (ENSG00000138942), score: -0.64 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.75 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.78 RPAINRPA interacting protein (ENSG00000129197), score: 0.72 SETD5SET domain containing 5 (ENSG00000168137), score: -0.74 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: 0.65 SPASTspastin (ENSG00000021574), score: 0.64 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.76 SPENspen homolog, transcriptional regulator (Drosophila) (ENSG00000065526), score: -0.74 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.71 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.86 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.63 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.94 STOML3stomatin (EPB72)-like 3 (ENSG00000133115), score: 0.63 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.65 TBCEtubulin folding cofactor E (ENSG00000116957), score: 0.66 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.78 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.66 TMEM33transmembrane protein 33 (ENSG00000109133), score: 0.64 TMEM60transmembrane protein 60 (ENSG00000135211), score: 0.71 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.67 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.65 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.76 TRNAU1APtRNA selenocysteine 1 associated protein 1 (ENSG00000180098), score: 0.7 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.72 TTC13tetratricopeptide repeat domain 13 (ENSG00000143643), score: 0.65 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.92 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.73 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.62 UBE4Aubiquitination factor E4A (UFD2 homolog, yeast) (ENSG00000110344), score: 0.69 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.69 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.64 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: -0.67 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.77 UTP23UTP23, small subunit (SSU) processome component, homolog (yeast) (ENSG00000147679), score: 0.67 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.98 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.77 WDR26WD repeat domain 26 (ENSG00000162923), score: -0.64 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.85 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.77 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.79 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.62 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.76 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.78 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.72 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.63 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.7
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_ts_m1_ca1 | oan | ts | m | 1 |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |
| oan_br_m_ca1 | oan | br | m | _ |
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_ts_m2_ca1 | oan | ts | m | 2 |
| oan_ts_m3_ca1 | oan | ts | m | 3 |
| oan_ht_m_ca1 | oan | ht | m | _ |