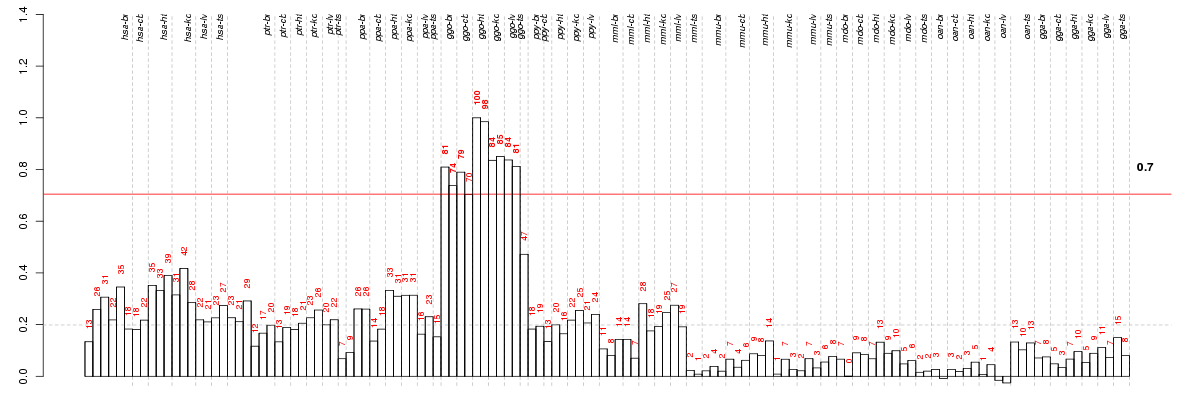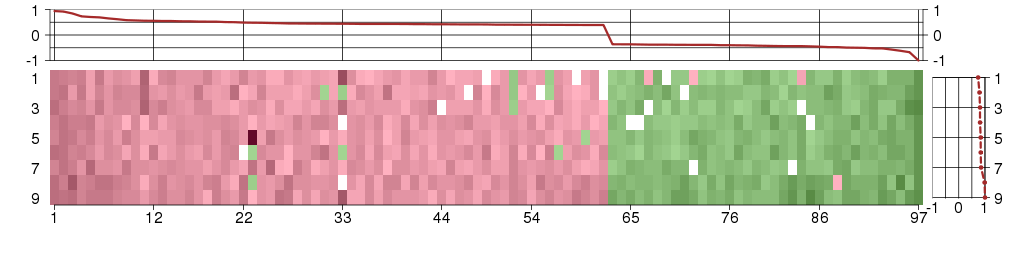

Under-expression is coded with green,
over-expression with red color.

mitotic cell cycle
Progression through the phases of the mitotic cell cycle, the most common eukaryotic cell cycle, which canonically comprises four successive phases called G1, S, G2, and M and includes replication of the genome and the subsequent segregation of chromosomes into daughter cells. In some variant cell cycles nuclear replication or nuclear division may not be followed by cell division, or G1 and G2 phases may be absent.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
protein ubiquitination
The process by which one or more ubiquitin moieties are added to a protein.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
cell cycle
The progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events. Canonically, the cell cycle comprises the replication and segregation of genetic material followed by the division of the cell, but in endocycles or syncytial cells nuclear replication or nuclear division may not be followed by cell division.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of protein ubiquitination
Any process that modulates the frequency, rate or extent of the addition of ubiquitin moieties to a protein.
regulation of protein modification process
Any process that modulates the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
protein modification by small protein conjugation
A protein modification process by which one or more moieties of a small protein, such as ubiquitin or a ubiquitin-like protein, are covalently attached to a target protein.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
macromolecule modification
The covalent alteration of one or more monomeric units in a polypeptide, polynucleotide, polysaccharide, or other biological macromolecule, resulting in a change in its properties.
post-translational protein modification
The covalent alteration of one or more amino acids occurring in a protein after the protein has been completely translated and released from the ribosome.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
regulation of ligase activity
Any process that modulates the frequency, rate or extent of ligase activity, the catalysis of the ligation of two substances with concomitant breaking of a diphosphate linkage, usually in a nucleoside triphosphate. Ligase is the systematic name for any enzyme of EC class 6.
regulation of ubiquitin-protein ligase activity
Any process that modulates the frequency, rate or extent of ubiquitin ligase activity, the catalysis of the reaction: ATP + ubiquitin + protein lysine = AMP + diphosphate + protein N-ubiquityllysine.
regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle
A cell cycle process that modulates the frequency, rate or extent of ubiquitin ligase activity that contributes to the mitotic cell cycle.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of molecular function
Any process that modulates the frequency, rate or extent of a molecular function, an elemental biological activity occurring at the molecular level, such as catalysis or binding.
protein modification by small protein conjugation or removal
A protein modification process by which one or more moieties of a small protein, such as ubiquitin or a ubiquitin-like protein, are covalently attached to or removed from a target protein.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
regulation of metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism.
regulation of cellular process
Any process that modulates the frequency, rate or extent of a cellular process, any of those that are carried out at the cellular level, but are not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of macromolecule metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
regulation of primary metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways within a cell or an organism involving those compounds formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cell cycle process
A cellular process that is involved in the progression of biochemical and morphological phases and events that occur in a cell during successive cell replication or nuclear replication events.
regulation of cellular metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways by which individual cells transform chemical substances.
regulation of catalytic activity
Any process that modulates the activity of an enzyme.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
regulation of protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle
A cell cycle process that modulates the frequency, rate or extent of ubiquitin ligase activity that contributes to the mitotic cell cycle.
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
protein modification process
The covalent alteration of one or more amino acids occurring in proteins, peptides and nascent polypeptides (co-translational, post-translational modifications). Includes the modification of charged tRNAs that are destined to occur in a protein (pre-translation modification).
regulation of cellular protein metabolic process
Any process that modulates the frequency, rate or extent of the chemical reactions and pathways involving a protein, occurring at the level of an individual cell.
regulation of protein modification process
Any process that modulates the frequency, rate or extent of the covalent alteration of one or more amino acid residues within a protein.
regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle
A cell cycle process that modulates the frequency, rate or extent of ubiquitin ligase activity that contributes to the mitotic cell cycle.
regulation of ubiquitin-protein ligase activity
Any process that modulates the frequency, rate or extent of ubiquitin ligase activity, the catalysis of the reaction: ATP + ubiquitin + protein lysine = AMP + diphosphate + protein N-ubiquityllysine.
regulation of protein ubiquitination
Any process that modulates the frequency, rate or extent of the addition of ubiquitin moieties to a protein.
protein ubiquitination
The process by which one or more ubiquitin moieties are added to a protein.


AAGABalpha- and gamma-adaptin binding protein (ENSG00000103591), score: -0.44 ABCC1ATP-binding cassette, sub-family C (CFTR/MRP), member 1 (ENSG00000103222), score: 0.43 AHCTF1AT hook containing transcription factor 1 (ENSG00000153207), score: -0.62 AHSA2AHA1, activator of heat shock 90kDa protein ATPase homolog 2 (yeast) (ENSG00000173209), score: -0.38 ANKRD28ankyrin repeat domain 28 (ENSG00000206560), score: -0.39 ANKRD54ankyrin repeat domain 54 (ENSG00000100124), score: -0.57 ARCN1archain 1 (ENSG00000095139), score: -0.47 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: 0.4 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: 0.43 BEST1bestrophin 1 (ENSG00000167995), score: 0.54 BUB1Bbudding uninhibited by benzimidazoles 1 homolog beta (yeast) (ENSG00000156970), score: 0.42 C12orf29chromosome 12 open reading frame 29 (ENSG00000133641), score: -0.39 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 0.94 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.92 CBFBcore-binding factor, beta subunit (ENSG00000067955), score: 0.46 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.45 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.54 CCDC90Bcoiled-coil domain containing 90B (ENSG00000137500), score: -0.37 CD79BCD79b molecule, immunoglobulin-associated beta (ENSG00000007312), score: 0.4 COG4component of oligomeric golgi complex 4 (ENSG00000103051), score: 0.42 COL8A1collagen, type VIII, alpha 1 (ENSG00000144810), score: 0.51 COLQcollagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase (ENSG00000206561), score: 0.43 COMMD10COMM domain containing 10 (ENSG00000145781), score: 0.45 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.62 CSF3Rcolony stimulating factor 3 receptor (granulocyte) (ENSG00000119535), score: 0.44 CUL1cullin 1 (ENSG00000055130), score: -1 CUL3cullin 3 (ENSG00000036257), score: -0.4 CWF19L1CWF19-like 1, cell cycle control (S. pombe) (ENSG00000095485), score: -0.39 CXCR4chemokine (C-X-C motif) receptor 4 (ENSG00000121966), score: 0.41 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: -0.42 DCTdopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (ENSG00000080166), score: 0.52 DDX55DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (ENSG00000111364), score: 0.5 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (ENSG00000101452), score: 0.42 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.46 DNTTdeoxynucleotidyltransferase, terminal (ENSG00000107447), score: 0.48 DVL3dishevelled, dsh homolog 3 (Drosophila) (ENSG00000161202), score: 0.55 EIF2B1eukaryotic translation initiation factor 2B, subunit 1 alpha, 26kDa (ENSG00000111361), score: 0.43 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.71 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: -0.53 FAM134Cfamily with sequence similarity 134, member C (ENSG00000141699), score: -0.41 FAM59Bfamily with sequence similarity 59, member B (ENSG00000157833), score: 0.65 FAM8A1family with sequence similarity 8, member A1 (ENSG00000137414), score: -0.37 GNALguanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type (ENSG00000141404), score: -0.44 HDDC2HD domain containing 2 (ENSG00000111906), score: 0.39 HKDC1hexokinase domain containing 1 (ENSG00000156510), score: 0.69 HMHA1histocompatibility (minor) HA-1 (ENSG00000180448), score: 0.43 IP6K3inositol hexakisphosphate kinase 3 (ENSG00000161896), score: 0.52 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.73 KIF15kinesin family member 15 (ENSG00000163808), score: 0.55 KLHDC10kelch domain containing 10 (ENSG00000128607), score: -0.36 KPNA6karyopherin alpha 6 (importin alpha 7) (ENSG00000025800), score: -0.43 LOC100289938similar to cytoplasmic tRNA 2-thiolation protein 2 (ENSG00000174177), score: 0.56 LUC7L2LUC7-like 2 (S. cerevisiae) (ENSG00000146963), score: 0.42 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: 0.57 MFNGMFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase (ENSG00000100060), score: 0.39 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.56 MRPS23mitochondrial ribosomal protein S23 (ENSG00000181610), score: 0.4 MYO1Gmyosin IG (ENSG00000136286), score: 0.4 MYT1myelin transcription factor 1 (ENSG00000196132), score: 0.41 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: -0.47 NEK6NIMA (never in mitosis gene a)-related kinase 6 (ENSG00000119408), score: 0.43 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: -0.52 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: 0.45 PCID2PCI domain containing 2 (ENSG00000126226), score: 0.48 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.49 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.58 PKP1plakophilin 1 (ectodermal dysplasia/skin fragility syndrome) (ENSG00000081277), score: 0.44 PLA2R1phospholipase A2 receptor 1, 180kDa (ENSG00000153246), score: 0.47 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.45 PPM1Mprotein phosphatase, Mg2+/Mn2+ dependent, 1M (ENSG00000164088), score: 0.4 PPPDE2PPPDE peptidase domain containing 2 (ENSG00000100418), score: -0.39 PRELPproline/arginine-rich end leucine-rich repeat protein (ENSG00000188783), score: 0.39 PSMB4proteasome (prosome, macropain) subunit, beta type, 4 (ENSG00000159377), score: 0.42 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: -0.38 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.84 QSER1glutamine and serine rich 1 (ENSG00000060749), score: -0.44 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: -0.67 RECQL4RecQ protein-like 4 (ENSG00000160957), score: 0.53 RNF111ring finger protein 111 (ENSG00000157450), score: -0.38 ROBO4roundabout homolog 4, magic roundabout (Drosophila) (ENSG00000154133), score: 0.39 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: 0.43 SMG7Smg-7 homolog, nonsense mediated mRNA decay factor (C. elegans) (ENSG00000116698), score: -0.41 SNX33sorting nexin 33 (ENSG00000173548), score: 0.44 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: 0.45 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: 0.41 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: -0.38 TMEM41Btransmembrane protein 41B (ENSG00000166471), score: -0.42 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: -0.49 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.44 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: 0.4 UBL3ubiquitin-like 3 (ENSG00000122042), score: -0.4 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (ENSG00000012963), score: -0.5 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: -0.37 WDR59WD repeat domain 59 (ENSG00000103091), score: 0.4 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: -0.5 XRCC3X-ray repair complementing defective repair in Chinese hamster cells 3 (ENSG00000126215), score: 0.43 ZFYVE21zinc finger, FYVE domain containing 21 (ENSG00000100711), score: 0.42
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |