Previous module |
Next module
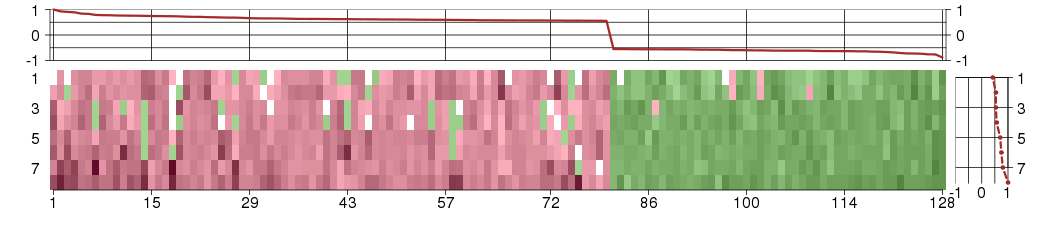
Module #316, TG: 2.4, TC: 2, 128 probes, 126 Entrez genes, 8 conditions


HELP
The image plot shows the color-coded level of gene expression, for the
genes and conditions in a given transcription module. The genes are on
the horizontal, the conditions on the vertical axis.
The genes are ordered according to their ISA gene scores, similarly
the conditions are ordered according to their condition scores. The
score of a gene means the «degree of inclusion» in
the module: a high score gene is essential in the module.
Condition scores can also be negative, that means that the genes of
the module are all down-regulated in the condition. Here the absolute
value of the score gives the «degree of inclusion».
The plots above and beside the expression matrix show the gene scores
and condition scores, respectively.
Note that the plot is interactive, you can see the name of the gene
and condition under the mouse cursor.
The expression matrix was normalized to have mean zero and standard
deviation one for every gene separately across all conditions
(i.e. not just for the conditions in the module).
— Click on the Help button again to close this help window.

Under-expression is coded with green,
over-expression with red color.
Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Biological processes
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for biological processes.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

response to tumor cell
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a tumor cell.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
all
NA
Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Cellular Components
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for cellular components.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
The GO tree — Molecular Function
HELP
This is one of three sections showing Gene Ontology enrichment of the
current module: in this case for molecular function.
The graph shows the hierarchy of the GO categories, their enrichment
for the current module is color coded, and the blue number beside the
category is the minus log ten p-value of the enrichment. (Calculated
using the standard hypergeometric test.) The color of the arrows code
«is a» (cyan) and «part of» relationships.
The tree was built the following way. First all GO terms with more
significant enrichment p-value than 0.05 were collected. Then all
paths from these terms to the root node of the GO tree were included
too. If a GO term is included more than once in the tree, then the
green numbers show 1) the id of the node, this makes it easier to find
other appereances of the term, and 2) the number of appearences.
Note that the same GO category might show up on the graph many
times. This is because the GO was «straightened» for this
graph, i.e. if there are more paths from a GO term to the root node of
the tree, all of them are included. The green numbers
Move the mouse cursor over the terms to get their definition. Clicking
on them takes you to the corresponding Gene Ontology web page.
If you cannot see a graph here at all, that means that there were no
significantly enriched GO categories, at the 0.05 level.
— Click on the Help button again to close this help window.

Help |
Hide |
Top
Help |
Show |
Top
GO BP test for over-representation
HELP
List of all enriched GO categories (biological processes), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0002347 | 7.783e-03 | 0.1098 | 3
CD40LG, IL12B, TP63 | 5 | response to tumor cell |
| GO:0032729 | 1.312e-02 | 0.1318 | 3
CD40LG, IL12B, IRF8 | 6 | positive regulation of interferon-gamma production |
| GO:0042108 | 1.583e-02 | 0.3294 | 4
CD28, EREG, IL12B, REL | 15 | positive regulation of cytokine biosynthetic process |
| GO:0030097 | 1.899e-02 | 2.701 | 10
ARNT, ATP7A, BPGM, CD28, CD40LG, IL12B, IRF8, PATZ1, TM7SF4, TNFRSF11A | 123 | hemopoiesis |
| GO:0032655 | 2.002e-02 | 0.1537 | 3
CD40LG, IRF8, REL | 7 | regulation of interleukin-12 production |
| GO:0021884 | 2.581e-02 | 0.04392 | 2
ATP7A, LHX8 | 2 | forebrain neuron development |
| GO:0048534 | 2.753e-02 | 2.877 | 10
ARNT, ATP7A, BPGM, CD28, CD40LG, IL12B, IRF8, PATZ1, TM7SF4, TNFRSF11A | 131 | hemopoietic or lymphoid organ development |
| GO:0032615 | 2.782e-02 | 0.1757 | 3
CD40LG, IRF8, REL | 8 | interleukin-12 production |
| GO:0001817 | 2.800e-02 | 1.449 | 7
ARNT, CD28, CD40LG, EREG, IL12B, IRF8, REL | 66 | regulation of cytokine production |
| GO:0001819 | 4.054e-02 | 0.7686 | 5
ARNT, CD40LG, EREG, IL12B, IRF8 | 35 | positive regulation of cytokine production |
| GO:0000122 | 4.057e-02 | 2.042 | 8
ETV3, IRF8, MBD3, NR0B1, ORC2L, PDX1, TP63, ZFHX3 | 93 | negative regulation of transcription from RNA polymerase II promoter |
| GO:0002521 | 4.130e-02 | 1.581 | 7
ATP7A, CD28, CD40LG, IL12B, PATZ1, TM7SF4, TNFRSF11A | 72 | leukocyte differentiation |
| GO:0002520 | 4.375e-02 | 3.118 | 10
ARNT, ATP7A, BPGM, CD28, CD40LG, IL12B, IRF8, PATZ1, TM7SF4, TNFRSF11A | 142 | immune system development |
| GO:0016070 | 4.643e-02 | 15.7 | 28
ADAT2, ARNT, CELF1, EREG, ESR2, ETV3, HMG20A, IKZF3, IRF8, LASS6, LHX8, LSM4, MBD3, NR0B1, ORC2L, PAIP1, PAPOLG, PARN, PATZ1, PDX1, PLRG1, POLR1B, QTRTD1, REL, SLBP, TMX1, TP63, ZFHX3 | 715 | RNA metabolic process |
| GO:0001816 | 4.643e-02 | 1.625 | 7
ARNT, CD28, CD40LG, EREG, IL12B, IRF8, REL | 74 | cytokine production |
| GO:0002831 | 4.724e-02 | 0.2196 | 3
CD28, CD40LG, IL12B | 10 | regulation of response to biotic stimulus |
Help |
Hide |
Top
Help |
Show |
Top
GO CC test for over-representation
HELP
List of all enriched GO categories (cellular components), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
GO MF test for over-representation
HELP
List of all enriched GO categories (molecular function), at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given GO term, just by chance.
- Count
is the number of genes in the module annotated with the given GO
term.
- Size is the total number of genes (in our universe)
annotated with the GO term.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
GO category.
Clicking on the GO identifiers takes you to the Gene Ontology web
pages.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| GO:0090079 | 4.904e-02 | 0.06799 | 2
CELF1, PAIP1 | 3 | translation regulator activity, nucleic acid binding |
Help |
Hide |
Top
Help |
Show |
Top
KEGG Pathway test for over-representation
HELP
List of all enriched KEGG pathways, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module annotated with the given KEGG pathway, just by chance.
- Count
is the number of genes in the module annotated with the given KEGG
pathway.
- Size is the total number of genes (in our universe)
annotated with the KEGG pathway.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
KEGG pathway.
Clicking on the KEGG identifiers takes you to the KEGG web site.
— Click on the Help button again to close this help window.
| Id | Pvalue | ExpCount | Count | Size | Term |
| 05330 | 9.517e-03 | 0.1297 | 3
CD28, CD40LG, IL12B | 5 | Allograft rejection |
| 04940 | 1.619e-02 | 0.1556 | 3
CD28, CPE, IL12B | 6 | Type I diabetes mellitus |
| 05320 | 3.279e-02 | 0.2075 | 3
CD28, CD40LG, TSHB | 8 | Autoimmune thyroid disease |
Help |
Hide |
Top
Help |
Show |
Top
miRNA test for over-representation
HELP
List of all enriched miRNA families, at the 0.05
p-value level.
The columns:
- ExpCount is the expected count of genes in the
module regulated by the given miRNA family, just by chance.
- Count
is the number of genes in the module regulated by the given miRNA
family.
- Size is the total number of genes (in our universe)
regulated with the given miRNA family.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
miRNA family.
The miRNA regulation data was taken from the TargetScan database.
(Only the conserved sites were used for the current analysis.)
Clicking on the miRNA names takes you to the TargetScan web site.
— Click on the Help button again to close this help window.
Help |
Hide |
Top
Help |
Show |
Top
Chromosome test for over-representation
HELP
List of all enriched Chromosomes, at the 0.05
p-value level.
The columns:
- ExpCount is the expected number of genes in the
module on the given chromosome, just by chance.
- Count
is the number of genes in the module on the given chromosome.
- Size is the total number of genes (in our universe)
on the given chromosome.
Clicking on Count shows the genes that drive the
enrichment. You can also click on the individual numbers in
the Count column, to show the driving genes for that individual
chromosome.
— Click on the Help button again to close this help window.
HELP
A list of all genes in the current module, in alphabetical order. The
size of the text corresponds to the gene scores.
Text color indicates correlation and anti-correlation: genes of the
same color are correlated, both move up or down in the module samples
(listed below). Genes of different color are anti-correlated, they
have opposite behavior in the module samples.
Note, that text color of the individual genes should be interpreted
together with the coloring of the samples below. For
red samples,
red genes have a higher expression
(compared to the average gene expression level),
green genes have a lower
expression.
Green samples have opposite
behavior, in these red genes have a
lower expression, green genes
have a higher expression.
Note that some gene symbols may show up more than once, if many
probes match the same Entrez gene.
Genes with no Entrez mapping are given separately, with their
Affymetrics probe ID.
— Click on the Help button again to close this help window.
Entrez genes
ABCA12ATP-binding cassette, sub-family A (ABC1), member 12 (ENSG00000144452), score: 0.75
ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.7
ADAT2adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) (ENSG00000189007), score: 0.61
AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.57
AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.57
APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.89
ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (ENSG00000102606), score: -0.56
ARL3ADP-ribosylation factor-like 3 (ENSG00000138175), score: -0.62
ARNTaryl hydrocarbon receptor nuclear translocator (ENSG00000143437), score: 0.63
ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.58
ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.61
BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: -0.6
BRD4bromodomain containing 4 (ENSG00000141867), score: 0.6
BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.61
C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.84
C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.65
C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: 0.62
C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.64
C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.63
CA6carbonic anhydrase VI (ENSG00000131686), score: 0.76
CAPN2calpain 2, (m/II) large subunit (ENSG00000162909), score: -0.56
CD28CD28 molecule (ENSG00000178562), score: 0.65
CD40LGCD40 ligand (ENSG00000102245), score: 0.73
CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 1
CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.63
CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.71
CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.67
CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.61
CPEcarboxypeptidase E (ENSG00000109472), score: -0.57
DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (ENSG00000169598), score: 0.62
DHX29DEAH (Asp-Glu-Ala-His) box polypeptide 29 (ENSG00000067248), score: -0.58
DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.72
EREGepiregulin (ENSG00000124882), score: 0.69
ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.6
ETV3ets variant 3 (ENSG00000117036), score: 0.63
FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.78
FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.77
FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.6
FNBP4formin binding protein 4 (ENSG00000109920), score: 0.6
FNDC7fibronectin type III domain containing 7 (ENSG00000143107), score: 0.57
GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: -0.55
HMG20Ahigh-mobility group 20A (ENSG00000140382), score: -0.56
HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.91
ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.76
IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.58
IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.79
IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.74
IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.74
INPP5Kinositol polyphosphate-5-phosphatase K (ENSG00000132376), score: -0.57
IRF8interferon regulatory factor 8 (ENSG00000140968), score: 0.65
KIAA0368KIAA0368 (ENSG00000136813), score: -0.56
KLHDC1kelch domain containing 1 (ENSG00000197776), score: 0.57
KLHL28kelch-like 28 (Drosophila) (ENSG00000179454), score: 0.58
KREMEN1kringle containing transmembrane protein 1 (ENSG00000183762), score: -0.61
LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.68
LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.6
LATS1LATS, large tumor suppressor, homolog 1 (Drosophila) (ENSG00000131023), score: 0.59
LCMT1leucine carboxyl methyltransferase 1 (ENSG00000205629), score: -0.71
LHX8LIM homeobox 8 (ENSG00000162624), score: 0.68
LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.65
LSM4LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) (ENSG00000130520), score: -0.63
MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.56
MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.57
MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.57
NR0B1nuclear receptor subfamily 0, group B, member 1 (ENSG00000169297), score: 0.56
OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.66
ORC2Lorigin recognition complex, subunit 2-like (yeast) (ENSG00000115942), score: -0.58
ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.64
PAIP1poly(A) binding protein interacting protein 1 (ENSG00000172239), score: -0.64
PANX3pannexin 3 (ENSG00000154143), score: 0.83
PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.73
PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (ENSG00000140694), score: -0.63
PATZ1POZ (BTB) and AT hook containing zinc finger 1 (ENSG00000100105), score: -0.62
PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.62
PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.65
PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.73
POF1Bpremature ovarian failure, 1B (ENSG00000124429), score: 0.56
POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.71
POLR1Bpolymerase (RNA) I polypeptide B, 128kDa (ENSG00000125630), score: 0.57
PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.7
QPCTLglutaminyl-peptide cyclotransferase-like (ENSG00000011478), score: 0.61
QTRTD1queuine tRNA-ribosyltransferase domain containing 1 (ENSG00000151576), score: 0.56
RAB19RAB19, member RAS oncogene family (ENSG00000146955), score: 0.56
RAB3GAP1RAB3 GTPase activating protein subunit 1 (catalytic) (ENSG00000115839), score: -0.61
RAB4ARAB4A, member RAS oncogene family (ENSG00000168118), score: -0.63
RBP1retinol binding protein 1, cellular (ENSG00000114115), score: -0.6
RELv-rel reticuloendotheliosis viral oncogene homolog (avian) (ENSG00000162924), score: 0.64
RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.62
RHOBTB3Rho-related BTB domain containing 3 (ENSG00000164292), score: -0.56
RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.76
SDF4stromal cell derived factor 4 (ENSG00000078808), score: -0.59
SEPT2septin 2 (ENSG00000168385), score: -0.76
SLBPstem-loop binding protein (ENSG00000163950), score: -0.59
SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.89
SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.93
SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (ENSG00000184402), score: -0.56
SSNA1Sjogren syndrome nuclear autoantigen 1 (ENSG00000176101), score: -0.56
SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.58
TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.59
TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.74
TATDN1TatD DNase domain containing 1 (ENSG00000147687), score: 0.57
TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.56
TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.72
TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.61
TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.63
TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.62
TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.66
TNFRSF11Atumor necrosis factor receptor superfamily, member 11a, NFKB activator (ENSG00000141655), score: 0.76
TP63tumor protein p63 (ENSG00000073282), score: 0.57
TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.59
TSHBthyroid stimulating hormone, beta (ENSG00000134200), score: 0.56
TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: -0.63
TUBB1tubulin, beta 1 (ENSG00000101162), score: 0.63
UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.55
UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.62
UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.61
UBL7ubiquitin-like 7 (bone marrow stromal cell-derived) (ENSG00000138629), score: -0.56
UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.67
VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.56
WDR41WD repeat domain 41 (ENSG00000164253), score: -0.76
XPR1xenotropic and polytropic retrovirus receptor 1 (ENSG00000143324), score: 0.62
ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.74
ZFHX3zinc finger homeobox 3 (ENSG00000140836), score: 0.59
ZMYM2zinc finger, MYM-type 2 (ENSG00000121741), score: -0.59
ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.68
ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.57
Non-Entrez genes
ENSG00000138175Unknown, score: -0.63
ENSG00000139921Unknown, score: 0.6
HELP
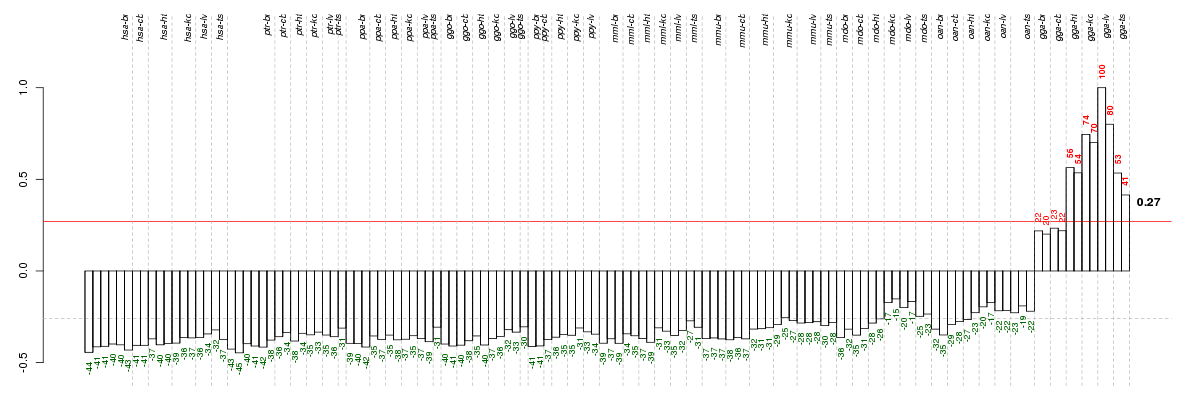
Conditions in the module, given in the same order as on the expression
plot above. Red color means over-expression, green under-expression in
the given condition.
The barplot below shows the condition (sample) scores. A separate bar
is shown for each sample, its height is the corresponding score of the
sample in the module. The red and green numbers on the bars are the
sample scores expressed in percents, i.e. 100% is 1.0.
The height of each bar corresponds to the weighted
mean expression of the module genes. The weights are the gene scores
of the module and they are positive for the genes listed in
red above and they are negative for
the genes that are listed in
green.
Bars going up correspond to samples listed in
red (the ones that are different
enough to included in the module). In these samples
the red module genes are highly
expressed, and the green
module genes are lowly expressed. The behavior of the genes is the
opposite for bars going down.
The red and green lines show the module thresholds, samples above
the red line and below the green line are included in the module.
The different species and tissues that were part of the study, are separated
by dashed vertical lines.
— Click on the Help button again to close this help window.
| Id | species | tissue | sex | individual |
| gga_ts_m2_ca1 | gga | ts | m | 2 |
| gga_ts_m1_ca1 | gga | ts | m | 1 |
| gga_ht_f_ca1 | gga | ht | f | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_lv_f_ca1 | gga | lv | f | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |



© 2008-2010 Computational Biology Group, Department of Medical Genetics,
University of Lausanne, Switzerland