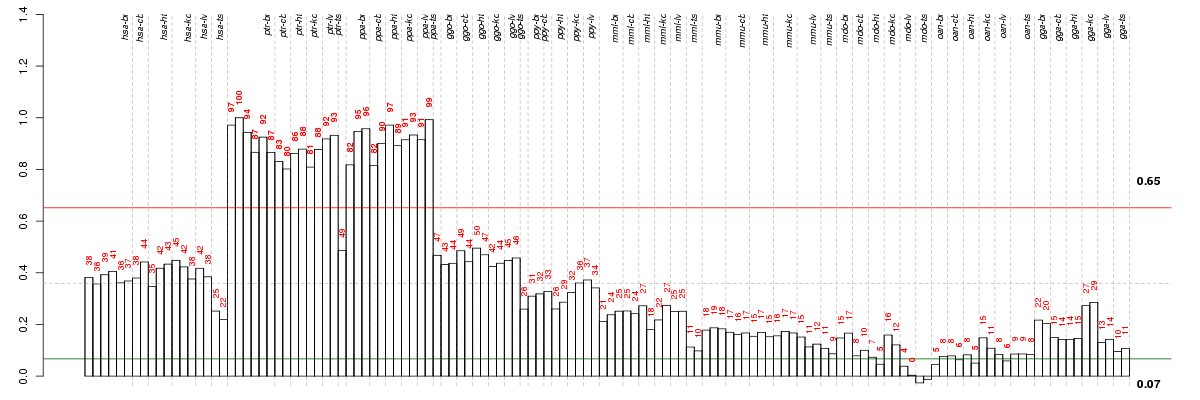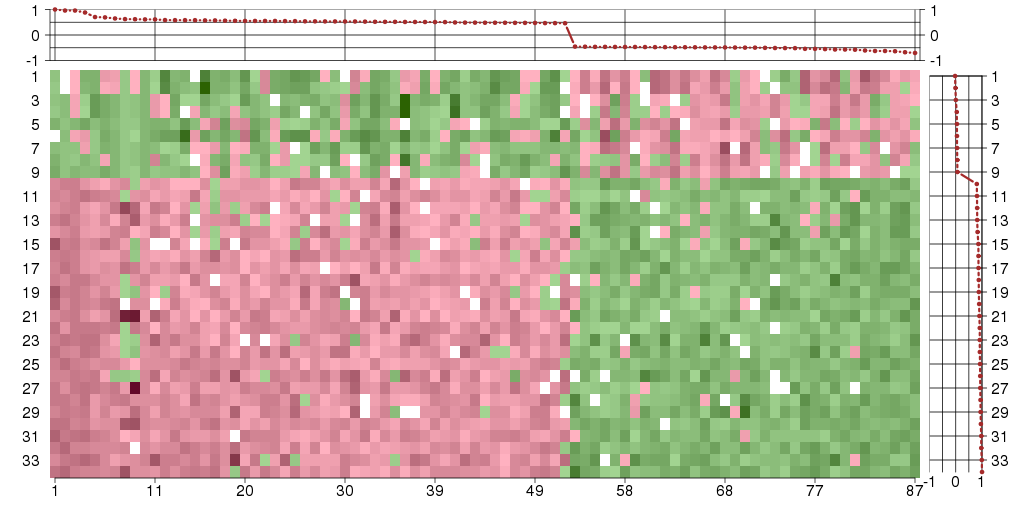

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
Golgi apparatus
A compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack. The Golgi apparatus differs from the endoplasmic reticulum in often having slightly thicker membranes, appearing in sections as a characteristic shallow semicircle so that the convex side (cis or entry face) abuts the endoplasmic reticulum, secretory vesicles emerging from the concave side (trans or exit face). In vertebrate cells there is usually one such organelle, while in invertebrates and plants, where they are known usually as dictyosomes, there may be several scattered in the cytoplasm. The Golgi apparatus processes proteins produced on the ribosomes of the rough endoplasmic reticulum; such processing includes modification of the core oligosaccharides of glycoproteins, and the sorting and packaging of proteins for transport to a variety of cellular locations. Three different regions of the Golgi are now recognized both in terms of structure and function: cis, in the vicinity of the cis face, trans, in the vicinity of the trans face, and medial, lying between the cis and trans regions.
cis-Golgi network
The network of interconnected tubular and cisternal structures located at the convex side of the Golgi apparatus, which abuts the endoplasmic reticulum.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
Golgi apparatus part
Any constituent part of the Golgi apparatus, a compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
Golgi apparatus
A compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack. The Golgi apparatus differs from the endoplasmic reticulum in often having slightly thicker membranes, appearing in sections as a characteristic shallow semicircle so that the convex side (cis or entry face) abuts the endoplasmic reticulum, secretory vesicles emerging from the concave side (trans or exit face). In vertebrate cells there is usually one such organelle, while in invertebrates and plants, where they are known usually as dictyosomes, there may be several scattered in the cytoplasm. The Golgi apparatus processes proteins produced on the ribosomes of the rough endoplasmic reticulum; such processing includes modification of the core oligosaccharides of glycoproteins, and the sorting and packaging of proteins for transport to a variety of cellular locations. Three different regions of the Golgi are now recognized both in terms of structure and function: cis, in the vicinity of the cis face, trans, in the vicinity of the trans face, and medial, lying between the cis and trans regions.
Golgi apparatus part
Any constituent part of the Golgi apparatus, a compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack.
Golgi apparatus part
Any constituent part of the Golgi apparatus, a compound membranous cytoplasmic organelle of eukaryotic cells, consisting of flattened, ribosome-free vesicles arranged in a more or less regular stack.

ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: 0.47 ATMINATM interactor (ENSG00000166454), score: 0.49 ATP6V1C2ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C2 (ENSG00000143882), score: 0.51 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: 0.48 BEST1bestrophin 1 (ENSG00000167995), score: 0.61 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: -0.48 C16orf73chromosome 16 open reading frame 73 (ENSG00000162039), score: 0.96 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: -0.55 C20orf118chromosome 20 open reading frame 118 (ENSG00000101342), score: 0.52 C7orf26chromosome 7 open reading frame 26 (ENSG00000146576), score: 0.54 C9orf167chromosome 9 open reading frame 167 (ENSG00000198113), score: 0.55 CCDC73coiled-coil domain containing 73 (ENSG00000186714), score: 0.51 CCM2cerebral cavernous malformation 2 (ENSG00000136280), score: 0.47 CELcarboxyl ester lipase (bile salt-stimulated lipase) (ENSG00000170835), score: 0.65 CNIH4cornichon homolog 4 (Drosophila) (ENSG00000143771), score: 0.46 CNOT6CCR4-NOT transcription complex, subunit 6 (ENSG00000113300), score: -0.54 CNTD1cyclin N-terminal domain containing 1 (ENSG00000176563), score: -0.63 COG3component of oligomeric golgi complex 3 (ENSG00000136152), score: 0.61 COL7A1collagen, type VII, alpha 1 (ENSG00000114270), score: 0.55 CSNK1G1casein kinase 1, gamma 1 (ENSG00000169118), score: -0.46 DDX17DEAD (Asp-Glu-Ala-Asp) box polypeptide 17 (ENSG00000100201), score: 0.48 DDX51DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 (ENSG00000185163), score: 0.54 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: 0.57 DNAJC10DnaJ (Hsp40) homolog, subfamily C, member 10 (ENSG00000077232), score: -0.45 DRG2developmentally regulated GTP binding protein 2 (ENSG00000108591), score: 0.58 DTX4deltex homolog 4 (Drosophila) (ENSG00000110042), score: 0.53 DUS4Ldihydrouridine synthase 4-like (S. cerevisiae) (ENSG00000105865), score: 0.54 ENTPD1ectonucleoside triphosphate diphosphohydrolase 1 (ENSG00000138185), score: 1 FAM101Bfamily with sequence similarity 101, member B (ENSG00000183688), score: 0.55 FAM134Cfamily with sequence similarity 134, member C (ENSG00000141699), score: 0.53 FBXO22F-box protein 22 (ENSG00000167196), score: -0.47 FKTNfukutin (ENSG00000106692), score: -0.48 GABPAGA binding protein transcription factor, alpha subunit 60kDa (ENSG00000154727), score: -0.5 GRTP1growth hormone regulated TBC protein 1 (ENSG00000139835), score: 0.7 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: -0.49 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: 0.58 HAGHhydroxyacylglutathione hydrolase (ENSG00000063854), score: 0.49 HEATR2HEAT repeat containing 2 (ENSG00000164818), score: -0.47 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: -0.58 HEXDChexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing (ENSG00000169660), score: 0.48 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: -0.61 HSCBHscB iron-sulfur cluster co-chaperone homolog (E. coli) (ENSG00000100209), score: -0.64 IMPG1interphotoreceptor matrix proteoglycan 1 (ENSG00000112706), score: 0.62 KANK3KN motif and ankyrin repeat domains 3 (ENSG00000186994), score: 0.88 KCNRGpotassium channel regulator (ENSG00000198553), score: 0.52 KIAA0146KIAA0146 (ENSG00000164808), score: 0.55 KIAA0174KIAA0174 (ENSG00000182149), score: 0.48 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: 0.51 LACE1lactation elevated 1 (ENSG00000135537), score: -0.5 LIG3ligase III, DNA, ATP-dependent (ENSG00000005156), score: -0.48 MAP3K15mitogen-activated protein kinase kinase kinase 15 (ENSG00000180815), score: 0.69 MKKSMcKusick-Kaufman syndrome (ENSG00000125863), score: 0.56 MRP63mitochondrial ribosomal protein 63 (ENSG00000173141), score: -0.68 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: -0.57 MTERFD3MTERF domain containing 3 (ENSG00000120832), score: 0.48 MTMR3myotubularin related protein 3 (ENSG00000100330), score: -0.48 NEIL1nei endonuclease VIII-like 1 (E. coli) (ENSG00000140398), score: 0.59 NINninein (GSK3B interacting protein) (ENSG00000100503), score: 0.48 NUDCD1NudC domain containing 1 (ENSG00000120526), score: -0.54 PARS2prolyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000162396), score: -0.51 PHF5APHD finger protein 5A (ENSG00000100410), score: -0.47 POLG2polymerase (DNA directed), gamma 2, accessory subunit (ENSG00000136480), score: 0.96 POLR1Cpolymerase (RNA) I polypeptide C, 30kDa (ENSG00000171453), score: -0.49 POLR3Bpolymerase (RNA) III (DNA directed) polypeptide B (ENSG00000013503), score: -0.57 PPIGpeptidylprolyl isomerase G (cyclophilin G) (ENSG00000138398), score: 0.52 PRDM15PR domain containing 15 (ENSG00000141956), score: -0.7 PRPF8PRP8 pre-mRNA processing factor 8 homolog (S. cerevisiae) (ENSG00000174231), score: -0.49 RAD9ARAD9 homolog A (S. pombe) (ENSG00000172613), score: -0.51 RBM6RNA binding motif protein 6 (ENSG00000004534), score: 0.55 RNF214ring finger protein 214 (ENSG00000167257), score: -0.63 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: -0.48 SAMSN1SAM domain, SH3 domain and nuclear localization signals 1 (ENSG00000155307), score: 0.56 SFPQsplicing factor proline/glutamine-rich (ENSG00000116560), score: 0.53 STX16syntaxin 16 (ENSG00000124222), score: 0.47 SYCP2Lsynaptonemal complex protein 2-like (ENSG00000153157), score: 0.62 TAF1DTATA box binding protein (TBP)-associated factor, RNA polymerase I, D, 41kDa (ENSG00000166012), score: 0.58 TCOF1Treacher Collins-Franceschetti syndrome 1 (ENSG00000070814), score: 0.53 TMEM64transmembrane protein 64 (ENSG00000180694), score: -0.5 TNFAIP8L3tumor necrosis factor, alpha-induced protein 8-like 3 (ENSG00000183578), score: 0.57 TRAPPC4trafficking protein particle complex 4 (ENSG00000196655), score: -0.47 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: 0.51 URB2URB2 ribosome biogenesis 2 homolog (S. cerevisiae) (ENSG00000135763), score: -0.52 USP8ubiquitin specific peptidase 8 (ENSG00000138592), score: -0.49 VGLL4vestigial like 4 (Drosophila) (ENSG00000144560), score: -0.47 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.51 WDSUB1WD repeat, sterile alpha motif and U-box domain containing 1 (ENSG00000196151), score: -0.47 ZDHHC21zinc finger, DHHC-type containing 21 (ENSG00000175893), score: 0.52
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| oan_br_m_ca1 | oan | br | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| ptr_cb_f_ca1 | ptr | cb | f | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| ppa_cb_m_ca1 | ppa | cb | m | _ |
| ppa_br_m_ca1 | ppa | br | m | _ |
| ptr_cb_m_ca1 | ptr | cb | m | _ |
| ptr_ht_m_ca1 | ptr | ht | m | _ |
| ptr_br_f_ca1 | ptr | br | f | _ |
| ptr_br_m4_ca1 | ptr | br | m | 4 |
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ptr_ht_f_ca1 | ptr | ht | f | _ |
| ppa_ht_f_ca1 | ppa | ht | f | _ |
| ppa_cb_f_ca1 | ppa | cb | f | _ |
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| ppa_lv_m_ca1 | ppa | lv | m | _ |
| ptr_lv_m_ca1 | ptr | lv | m | _ |
| ptr_br_m5_ca1 | ptr | br | m | 5 |
| ptr_lv_f_ca1 | ptr | lv | f | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ppa_br_f2_ca1 | ppa | br | f | 2 |
| ppa_ht_m_ca1 | ppa | ht | m | _ |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ppa_lv_f_ca1 | ppa | lv | f | _ |
| ptr_br_m2_ca1 | ptr | br | m | 2 |