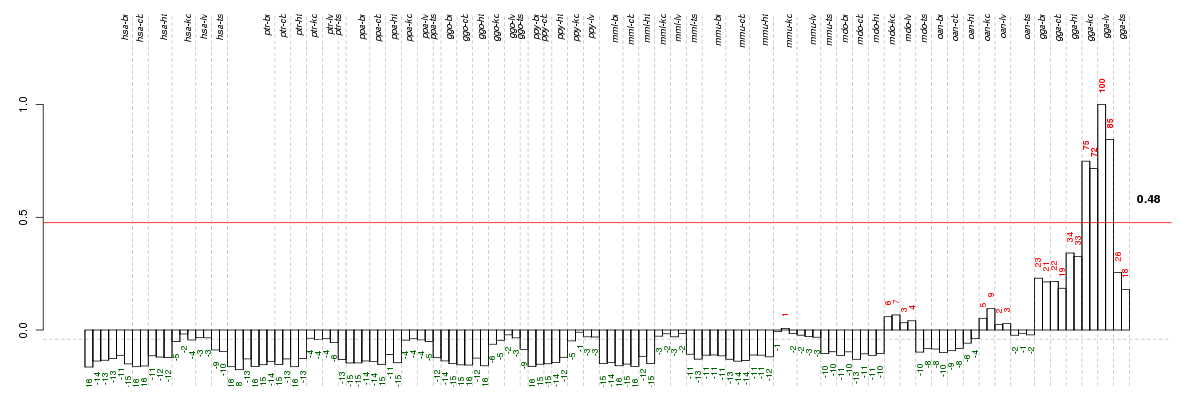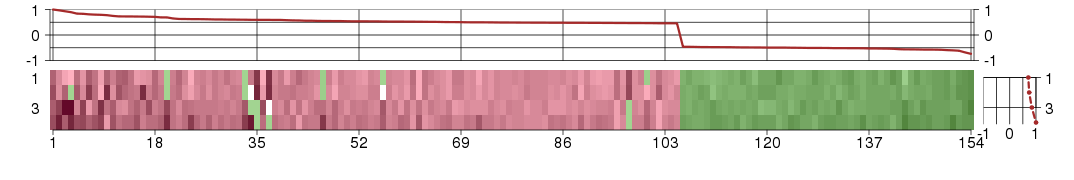

Under-expression is coded with green,
over-expression with red color.

cell activation
A change in the morphology or behavior of a cell resulting from exposure to an activating factor such as a cellular or soluble ligand.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
regulation of response to biotic stimulus
Any process that modulates the frequency, rate, or extent of a response to biotic stimulus.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
lymph node development
The process whose specific outcome is the progression of lymph nodes over time, from their formation to the mature structure. A lymph node is a round, oval, or bean shaped structure localized in clusters along the lymphatic vessels, with a distinct internal structure including specialized vasculature and B- and T-zones for the activation of lymphocytes.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
response to biotic stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a biotic stimulus, a stimulus caused or produced by a living organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
hemopoiesis
The process whose specific outcome is the progression of the myeloid and lymphoid derived organ/tissue systems of the blood and other parts of the body over time, from formation to the mature structure. The site of hemopoiesis is variable during development, but occurs primarily in bone marrow or kidney in many adult vertebrates.
lymphocyte differentiation
The process whereby a relatively unspecialized precursor cell acquires specialized features of B cells, T cells, or natural killer cells.
cell differentiation
The process whereby relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state.
T cell differentiation
The process by which a precursor cell type acquires characteristics of a more mature T-cell. A T cell is a type of lymphocyte whose definin characteristic is the expression of a T cell receptor complex.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
T cell activation
The change in morphology and behavior of a mature or immature T cell resulting from exposure to a mitogen, cytokine, chemokine, cellular ligand, or an antigen for which it is specific.
leukocyte activation
A change in morphology and behavior of a leukocyte resulting from exposure to a specific antigen, mitogen, cytokine, cellular ligand, or soluble factor.
lymphocyte activation
A change in morphology and behavior of a lymphocyte resulting from exposure to a specific antigen, mitogen, cytokine, chemokine, cellular ligand, or soluble factor.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
cellular developmental process
A biological process whose specific outcome is the progression of a cell over time from an initial condition to a later condition.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
leukocyte activation
A change in morphology and behavior of a leukocyte resulting from exposure to a specific antigen, mitogen, cytokine, cellular ligand, or soluble factor.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
regulation of response to biotic stimulus
Any process that modulates the frequency, rate, or extent of a response to biotic stimulus.
immune system development
The process whose specific outcome is the progression of an organismal system whose objective is to provide calibrated responses by an organism to a potential internal or invasive threat, over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
hemopoietic or lymphoid organ development
The process whose specific outcome is the progression of any organ involved in hemopoiesis or lymphoid cell activation over time, from its formation to the mature structure. Such development includes differentiation of resident cell types (stromal cells) and of migratory cell types dependent on the unique microenvironment afforded by the organ for their proper differentiation.
leukocyte differentiation
The process whereby a relatively unspecialized hemopoietic precursor cell acquires the specialized features of a plasmacytoid dendritic cell or any cell of the myeloid leukocyte or lymphocyte lineages.
T cell differentiation
The process by which a precursor cell type acquires characteristics of a more mature T-cell. A T cell is a type of lymphocyte whose definin characteristic is the expression of a T cell receptor complex.
lymphocyte differentiation
The process whereby a relatively unspecialized precursor cell acquires specialized features of B cells, T cells, or natural killer cells.


| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 05330 | 1.634e-02 | 0.1628 | 3 | 5 | Allograft rejection |
| 04672 | 3.151e-02 | 0.456 | 4 | 14 | Intestinal immune network for IgA production |
ABCA12ATP-binding cassette, sub-family A (ABC1), member 12 (ENSG00000144452), score: 0.69 ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.55 ADSLadenylosuccinate lyase (ENSG00000239900), score: 0.52 AHRaryl hydrocarbon receptor (ENSG00000106546), score: 0.48 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.53 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.63 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.51 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.68 ARHGEF7Rho guanine nucleotide exchange factor (GEF) 7 (ENSG00000102606), score: -0.52 ARL3ADP-ribosylation factor-like 3 (ENSG00000138175), score: -0.5 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.49 ARRDC3arrestin domain containing 3 (ENSG00000113369), score: 0.46 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.46 BAMBIBMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (ENSG00000095739), score: 0.59 BCMO1beta-carotene 15,15'-monooxygenase 1 (ENSG00000135697), score: 0.47 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: -0.56 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.48 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.6 C16orf57chromosome 16 open reading frame 57 (ENSG00000103005), score: 0.48 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.62 C20orf79chromosome 20 open reading frame 79 (ENSG00000132631), score: 0.53 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.73 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.8 CD28CD28 molecule (ENSG00000178562), score: 0.72 CD40LGCD40 ligand (ENSG00000102245), score: 0.73 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.84 CDH2cadherin 2, type 1, N-cadherin (neuronal) (ENSG00000170558), score: -0.48 CELF1CUGBP, Elav-like family member 1 (ENSG00000149187), score: -0.53 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.55 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.53 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.46 CLDN1claudin 1 (ENSG00000163347), score: 0.46 COCHcoagulation factor C homolog, cochlin (Limulus polyphemus) (ENSG00000100473), score: 0.55 CRISP1cysteine-rich secretory protein 1 (ENSG00000124812), score: 0.59 CYP7B1cytochrome P450, family 7, subfamily B, polypeptide 1 (ENSG00000172817), score: 0.49 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.49 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.48 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (ENSG00000169598), score: 0.48 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.56 DNMBPdynamin binding protein (ENSG00000107554), score: 0.54 EREGepiregulin (ENSG00000124882), score: 0.81 ESR2estrogen receptor 2 (ER beta) (ENSG00000140009), score: 0.71 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.56 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.65 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.55 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.48 FNBP4formin binding protein 4 (ENSG00000109920), score: 0.47 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.46 GEMIN4gem (nuclear organelle) associated protein 4 (ENSG00000179409), score: 0.47 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.49 HOXA13homeobox A13 (ENSG00000106031), score: 0.47 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 1 HSPA5heat shock 70kDa protein 5 (glucose-regulated protein, 78kDa) (ENSG00000044574), score: 0.47 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.72 IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.6 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.93 IL15interleukin 15 (ENSG00000164136), score: 0.47 IL1RAPinterleukin 1 receptor accessory protein (ENSG00000196083), score: 0.52 IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.9 IL7Rinterleukin 7 receptor (ENSG00000168685), score: 0.58 INTS2integrator complex subunit 2 (ENSG00000108506), score: 0.49 IRF8interferon regulatory factor 8 (ENSG00000140968), score: 0.62 KIAA1958KIAA1958 (ENSG00000165185), score: 0.47 KIF12kinesin family member 12 (ENSG00000136883), score: 0.51 KLHL14kelch-like 14 (Drosophila) (ENSG00000197705), score: 0.52 KRTCAP3keratinocyte associated protein 3 (ENSG00000157992), score: 0.48 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.59 LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.58 LCMT1leucine carboxyl methyltransferase 1 (ENSG00000205629), score: -0.6 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.79 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.51 LSM4LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) (ENSG00000130520), score: -0.47 MAP3K13mitogen-activated protein kinase kinase kinase 13 (ENSG00000073803), score: 0.54 MFI2antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 (ENSG00000163975), score: 0.62 MFSD10major facilitator superfamily domain containing 10 (ENSG00000109736), score: -0.47 NFKBIDnuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, delta (ENSG00000167604), score: 0.48 NLRC3NLR family, CARD domain containing 3 (ENSG00000167984), score: 0.57 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.51 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.49 NTSR1neurotensin receptor 1 (high affinity) (ENSG00000101188), score: 0.53 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.5 ORC2Lorigin recognition complex, subunit 2-like (yeast) (ENSG00000115942), score: -0.53 ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.57 OSTalphaorganic solute transporter alpha (ENSG00000163959), score: 0.54 PAIP1poly(A) binding protein interacting protein 1 (ENSG00000172239), score: -0.52 PANX3pannexin 3 (ENSG00000154143), score: 0.6 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.57 PATZ1POZ (BTB) and AT hook containing zinc finger 1 (ENSG00000100105), score: -0.62 PDE6Dphosphodiesterase 6D, cGMP-specific, rod, delta (ENSG00000156973), score: -0.56 PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.78 PICALMphosphatidylinositol binding clathrin assembly protein (ENSG00000073921), score: -0.52 PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.61 PLRG1pleiotropic regulator 1 (PRL1 homolog, Arabidopsis) (ENSG00000171566), score: -0.49 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.53 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.48 QPCTLglutaminyl-peptide cyclotransferase-like (ENSG00000011478), score: 0.53 RAB4ARAB4A, member RAS oncogene family (ENSG00000168118), score: -0.57 RBP1retinol binding protein 1, cellular (ENSG00000114115), score: -0.49 RGNregucalcin (senescence marker protein-30) (ENSG00000130988), score: 0.48 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.46 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.75 RRM1ribonucleotide reductase M1 (ENSG00000167325), score: -0.57 RUSC2RUN and SH3 domain containing 2 (ENSG00000198853), score: -0.49 RXFP3relaxin/insulin-like family peptide receptor 3 (ENSG00000182631), score: 0.59 SASH3SAM and SH3 domain containing 3 (ENSG00000122122), score: 0.5 SEPT2septin 2 (ENSG00000168385), score: -0.5 SESN1sestrin 1 (ENSG00000080546), score: 0.48 SGCEsarcoglycan, epsilon (ENSG00000127990), score: -0.49 SGK3serum/glucocorticoid regulated kinase family, member 3 (ENSG00000104205), score: 0.49 SHISA2shisa homolog 2 (Xenopus laevis) (ENSG00000180730), score: 0.51 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: 0.48 SLBPstem-loop binding protein (ENSG00000163950), score: -0.52 SLC24A6solute carrier family 24 (sodium/potassium/calcium exchanger), member 6 (ENSG00000089060), score: 0.61 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: -0.48 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.83 SNX33sorting nexin 33 (ENSG00000173548), score: 0.48 SOS1son of sevenless homolog 1 (Drosophila) (ENSG00000115904), score: 0.48 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.97 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (ENSG00000184402), score: -0.51 SSNA1Sjogren syndrome nuclear autoantigen 1 (ENSG00000176101), score: -0.46 SSR1signal sequence receptor, alpha (ENSG00000124783), score: 0.54 STOML3stomatin (EPB72)-like 3 (ENSG00000133115), score: 0.6 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.62 TADA2Btranscriptional adaptor 2B (ENSG00000173011), score: -0.47 TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.51 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.72 TMED8transmembrane emp24 protein transport domain containing 8 (ENSG00000100580), score: 0.47 TMEM199transmembrane protein 199 (ENSG00000244045), score: 0.5 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.56 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.49 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.73 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.53 TNFRSF11Atumor necrosis factor receptor superfamily, member 11a, NFKB activator (ENSG00000141655), score: 0.69 TRAF3IP3TRAF3 interacting protein 3 (ENSG00000009790), score: 0.51 TRAT1T cell receptor associated transmembrane adaptor 1 (ENSG00000163519), score: 0.52 TRPA1transient receptor potential cation channel, subfamily A, member 1 (ENSG00000104321), score: 0.55 TSHBthyroid stimulating hormone, beta (ENSG00000134200), score: 0.59 TTLL1tubulin tyrosine ligase-like family, member 1 (ENSG00000100271), score: -0.51 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.5 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.49 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.51 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.48 WDFY4WDFY family member 4 (ENSG00000128815), score: 0.57 WDR41WD repeat domain 41 (ENSG00000164253), score: -0.74 WNT4wingless-type MMTV integration site family, member 4 (ENSG00000162552), score: 0.5 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.5 XPR1xenotropic and polytropic retrovirus receptor 1 (ENSG00000143324), score: 0.49 YPEL5yippee-like 5 (Drosophila) (ENSG00000119801), score: -0.49 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.59 ZFAND5zinc finger, AN1-type domain 5 (ENSG00000107372), score: -0.54 ZMYM2zinc finger, MYM-type 2 (ENSG00000121741), score: -0.52 ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.63 ZP1zona pellucida glycoprotein 1 (sperm receptor) (ENSG00000149506), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_kd_f_ca1 | gga | kd | f | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_lv_f_ca1 | gga | lv | f | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |