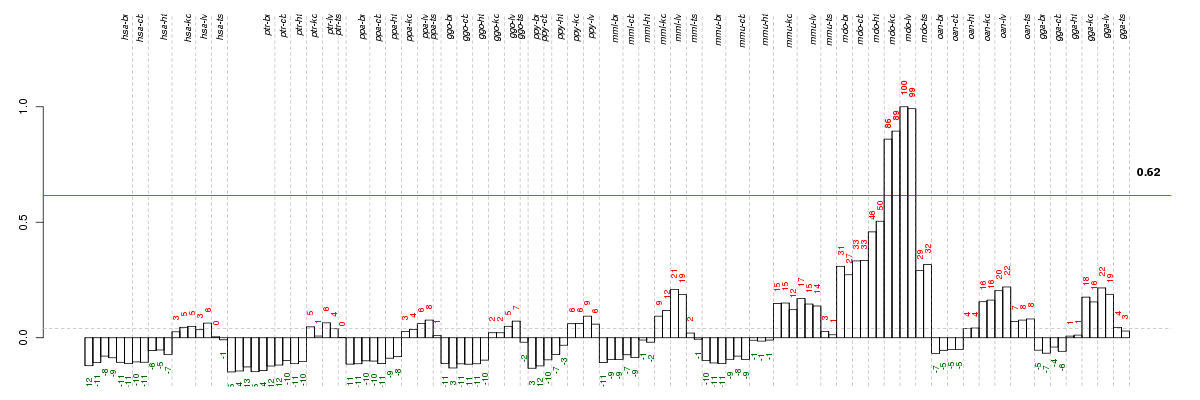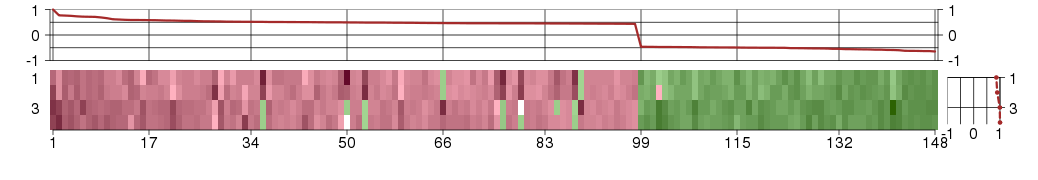

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.

AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.5 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (ENSG00000154265), score: -0.47 ABCG1ATP-binding cassette, sub-family G (WHITE), member 1 (ENSG00000160179), score: -0.5 ABHD6abhydrolase domain containing 6 (ENSG00000163686), score: -0.57 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.52 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.49 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.62 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.54 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.44 AIFM2apoptosis-inducing factor, mitochondrion-associated, 2 (ENSG00000042286), score: 0.46 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.52 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.56 ANKRD22ankyrin repeat domain 22 (ENSG00000152766), score: 0.46 AP2B1adaptor-related protein complex 2, beta 1 subunit (ENSG00000006125), score: -0.49 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.44 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.5 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 1 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.57 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.47 ATMINATM interactor (ENSG00000166454), score: -0.58 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.51 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.55 ATP2C1ATPase, Ca++ transporting, type 2C, member 1 (ENSG00000017260), score: -0.48 ATXN2ataxin 2 (ENSG00000204842), score: 0.5 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.46 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.48 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.45 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.48 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.53 CASS4Cas scaffolding protein family member 4 (ENSG00000087589), score: 0.46 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.57 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.52 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.51 CLDN19claudin 19 (ENSG00000164007), score: 0.45 CLTBclathrin, light chain B (ENSG00000175416), score: -0.63 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.56 CREB3L3cAMP responsive element binding protein 3-like 3 (ENSG00000060566), score: 0.52 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.52 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.55 CYTBcytochrome b (ENSG00000198727), score: -0.65 CYTH1cytohesin 1 (ENSG00000108669), score: -0.56 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.5 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.6 DHDPSLdihydrodipicolinate synthase-like, mitochondrial (ENSG00000241935), score: 0.44 DHRS7Bdehydrogenase/reductase (SDR family) member 7B (ENSG00000109016), score: -0.5 EEA1early endosome antigen 1 (ENSG00000102189), score: 0.56 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: 0.51 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.5 EPHA1EPH receptor A1 (ENSG00000146904), score: 0.46 EPYCepiphycan (ENSG00000083782), score: 0.53 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.47 FAM18Afamily with sequence similarity 18, member A (ENSG00000166676), score: -0.49 FAM3Dfamily with sequence similarity 3, member D (ENSG00000198643), score: 0.75 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.66 FERMT1fermitin family member 1 (ENSG00000101311), score: 0.51 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.48 GABRPgamma-aminobutyric acid (GABA) A receptor, pi (ENSG00000094755), score: 0.49 GCM1glial cells missing homolog 1 (Drosophila) (ENSG00000137270), score: 0.46 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.46 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.61 GRAMD2GRAM domain containing 2 (ENSG00000175318), score: 0.45 GSCgoosecoid homeobox (ENSG00000133937), score: 0.49 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.53 IFT46intraflagellar transport 46 homolog (Chlamydomonas) (ENSG00000118096), score: -0.47 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.52 IMPAD1inositol monophosphatase domain containing 1 (ENSG00000104331), score: -0.59 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.46 IQGAP1IQ motif containing GTPase activating protein 1 (ENSG00000140575), score: 0.44 KIAA1432KIAA1432 (ENSG00000107036), score: 0.45 KIFAP3kinesin-associated protein 3 (ENSG00000075945), score: -0.49 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.47 LYSMD3LysM, putative peptidoglycan-binding, domain containing 3 (ENSG00000176018), score: 0.46 MARVELD3MARVEL domain containing 3 (ENSG00000140832), score: 0.5 MPP1membrane protein, palmitoylated 1, 55kDa (ENSG00000130830), score: 0.54 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.58 MTMR14myotubularin related protein 14 (ENSG00000163719), score: 0.71 MYH10myosin, heavy chain 10, non-muscle (ENSG00000133026), score: -0.53 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.49 NADSYN1NAD synthetase 1 (ENSG00000172890), score: 0.54 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.49 ND2MTND2 (ENSG00000198763), score: -0.62 NDST4N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 4 (ENSG00000138653), score: 0.5 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.48 NOD1nucleotide-binding oligomerization domain containing 1 (ENSG00000106100), score: 0.47 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.71 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.51 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.49 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.46 OSBPL3oxysterol binding protein-like 3 (ENSG00000070882), score: 0.59 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.52 PDP2pyruvate dehyrogenase phosphatase catalytic subunit 2 (ENSG00000172840), score: 0.59 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.47 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.46 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.54 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.45 PODNpodocan (ENSG00000174348), score: 0.62 POLR2Cpolymerase (RNA) II (DNA directed) polypeptide C, 33kDa (ENSG00000102978), score: -0.46 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.77 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.48 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.47 QSER1glutamine and serine rich 1 (ENSG00000060749), score: 0.48 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.52 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.5 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.48 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.45 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.44 SDC2syndecan 2 (ENSG00000169439), score: 0.46 SFXN3sideroflexin 3 (ENSG00000107819), score: -0.47 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.5 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.49 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.52 SLC25A40solute carrier family 25, member 40 (ENSG00000075303), score: 0.51 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.69 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.57 SNX2sorting nexin 2 (ENSG00000205302), score: 0.58 SPATS2spermatogenesis associated, serine-rich 2 (ENSG00000123352), score: -0.5 STAB2stabilin 2 (ENSG00000136011), score: 0.52 STAMBPSTAM binding protein (ENSG00000124356), score: 0.5 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.59 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.57 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.47 TEX2testis expressed 2 (ENSG00000136478), score: 0.46 TEX264testis expressed 264 (ENSG00000164081), score: 0.49 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.44 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.73 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.48 TMEM53transmembrane protein 53 (ENSG00000126106), score: 0.46 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.47 TNMDtenomodulin (ENSG00000000005), score: 0.47 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.5 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.55 TSNtranslin (ENSG00000211460), score: -0.49 TTF2transcription termination factor, RNA polymerase II (ENSG00000116830), score: 0.76 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.59 TXNDC12thioredoxin domain containing 12 (endoplasmic reticulum) (ENSG00000117862), score: 0.6 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.63 UPK3Auroplakin 3A (ENSG00000100373), score: 0.45 VPS39vacuolar protein sorting 39 homolog (S. cerevisiae) (ENSG00000166887), score: -0.47 VPS54vacuolar protein sorting 54 homolog (S. cerevisiae) (ENSG00000143952), score: 0.46 WDR37WD repeat domain 37 (ENSG00000047056), score: -0.53 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.47 XCR1chemokine (C motif) receptor 1 (ENSG00000173578), score: 0.45 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -0.62 ZDHHC13zinc finger, DHHC-type containing 13 (ENSG00000177054), score: 0.72 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.45 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.54 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.45 ZPLD1zona pellucida-like domain containing 1 (ENSG00000170044), score: 0.51
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |