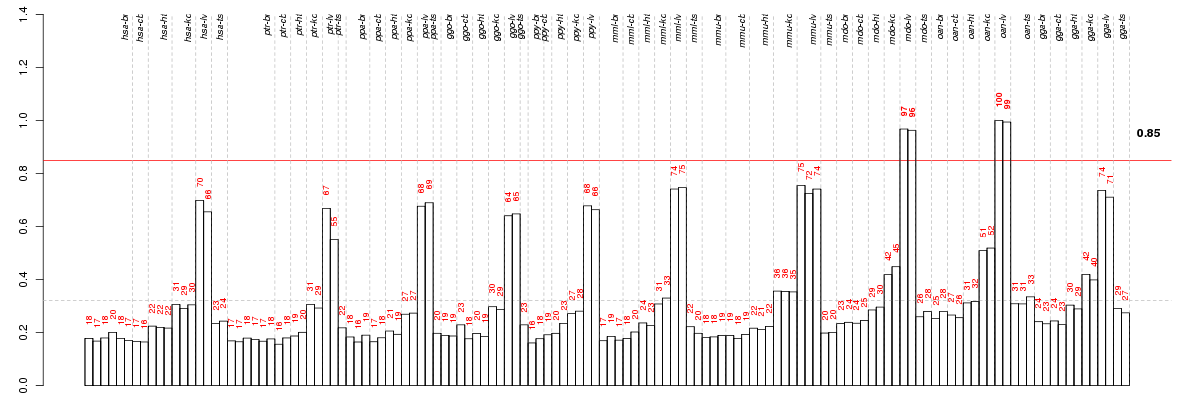

Under-expression is coded with green,
over-expression with red color.

sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
sulfur metabolic process
The chemical reactions and pathways involving the nonmetallic element sulfur or compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
adaptive immune response
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process, and allowing for enhanced response to subsequent exposures to the same antigen (immunological memory).
immune effector process
Any process of the immune system that occurs as part of an immune response.
activation of immune response
Any process that initiates an immune response.
immune system process
Any process involved in the development or functioning of the immune system, an organismal system for calibrated responses to potential internal or invasive threats.
leukocyte mediated immunity
Any process involved in the carrying out of an immune response by a leukocyte.
lymphocyte mediated immunity
Any process involved in the carrying out of an immune response by a lymphocyte.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
adaptive immune response based on somatic recombination of immune receptors built from immunoglobulin superfamily domains
An immune response based on directed amplification of specific receptors for antigen produced through a somatic diversification process that includes somatic recombination of germline gene segments encoding immunoglobulin superfamily domains, and allowing for enhanced responses upon subsequent exposures to the same antigen (immunological memory). Recombined receptors for antigen encoded by immunoglobulin superfamily domains include T cell receptors and immunoglobulins (antibodies). An example of this is the adaptive immune response found in Mus musculus.
acute inflammatory response
Inflammation which comprises a rapid, short-lived, relatively uniform response to acute injury or antigenic challenge and is characterized by accumulations of fluid, plasma proteins, and granulocytic leukocytes. An acute inflammatory response occurs within a matter of minutes or hours, and either resolves within a few days or becomes a chronic inflammatory response.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
defense response
Reactions, triggered in response to the presence of a foreign body or the occurrence of an injury, which result in restriction of damage to the organism attacked or prevention/recovery from the infection caused by the attack.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
protein maturation by peptide bond cleavage
The hydrolysis of a peptide bond or bonds within a protein as part of protein maturation, the process leading to the attainment of the full functional capacity of a protein.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.
glutamate metabolic process
The chemical reactions and pathways involving glutamate, the anion of 2-aminopentanedioic acid.
lipid metabolic process
The chemical reactions and pathways involving lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. Includes fatty acids; neutral fats, other fatty-acid esters, and soaps; long-chain (fatty) alcohols and waxes; sphingoids and other long-chain bases; glycolipids, phospholipids and sphingolipids; and carotenes, polyprenols, sterols, terpenes and other isoprenoids.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
lipid transport
The directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore. Lipids are compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
response to stress
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a disturbance in organismal or cellular homeostasis, usually, but not necessarily, exogenous (e.g. temperature, humidity, ionizing radiation).
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
humoral immune response
An immune response mediated through a body fluid.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
steroid metabolic process
The chemical reactions and pathways involving steroids, compounds with a 1,2,cyclopentanoperhydrophenanthrene nucleus.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
androgen metabolic process
The chemical reactions and pathways involving androgens, C19 steroid hormones that can stimulate the development of male sexual characteristics.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
biosynthetic process
The chemical reactions and pathways resulting in the formation of substances; typically the energy-requiring part of metabolism in which simpler substances are transformed into more complex ones.
glutamine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the glutamine family, comprising arginine, glutamate, glutamine and proline.
serine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the serine family, comprising cysteine, glycine, homoserine, selenocysteine and serine.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
amine biosynthetic process
The chemical reactions and pathways resulting in the formation of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
response to wounding
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
regulation of hormone levels
Any process that modulates the levels of hormone within an organism or a tissue. A hormone is any substance formed in very small amounts in one specialized organ or group of cells and carried (sometimes in the bloodstream) to another organ or group of cells in the same organism, upon which it has a specific regulatory action.
lipid localization
Any process by which a lipid is transported to, or maintained in, a specific location.
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
immunoglobulin mediated immune response
An immune response mediated by immunoglobulins, whether cell-bound or in solution.
protein processing
Any protein maturation process achieved by the cleavage of peptide bonds.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
monocarboxylic acid metabolic process
The chemical reactions and pathways involving monocarboxylic acids, any organic acid containing one carboxyl (COOH) group or anion (COO-).
macromolecule localization
Any process by which a macromolecule is transported to, or maintained in, a specific location.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
carboxylic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
response to stimulus
A change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
regulation of immune system process
Any process that modulates the frequency, rate, or extent of an immune system process.
positive regulation of biological process
Any process that activates or increases the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
immune response
Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat.
regulation of response to stimulus
Any process that modulates the frequency, rate or extent of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular biosynthetic process
The chemical reactions and pathways resulting in the formation of substances, carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
small molecule catabolic process
The chemical reactions and pathways resulting in the breakdown of small molecules, any monomeric molecule of small relative molecular mass.
small molecule biosynthetic process
The chemical reactions and pathways resulting in the formation of small molecules, any monomeric molecule of small relative molecular mass.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
positive regulation of immune system process
Any process that activates or increases the frequency, rate, or extent of an immune system process.
immune effector process
Any process of the immune system that occurs as part of an immune response.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
positive regulation of response to stimulus
Any process that activates, maintains or increases the rate of a response to a stimulus. Response to stimulus is a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus.
regulation of immune response
Any process that modulates the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
cellular nitrogen compound biosynthetic process
The chemical reactions and pathways resulting in the formation of organic and inorganic nitrogenous compounds.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
protein maturation
Any process leading to the attainment of the full functional capacity of a protein.
organic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of organic acids, any acidic compound containing carbon in covalent linkage.
organic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of organic acids, any acidic compound containing carbon in covalent linkage.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
lipid catabolic process
The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
positive regulation of immune response
Any process that activates or increases the frequency, rate or extent of the immune response, the immunological reaction of an organism to an immunogenic stimulus.
activation of immune response
Any process that initiates an immune response.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.
inflammatory response
The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages.
lipid transport
The directed movement of lipids into, out of, within or between cells by means of some external agent such as a transporter or pore. Lipids are compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent.
hormone metabolic process
The chemical reactions and pathways involving any hormone, naturally occurring substances secreted by specialized cells that affects the metabolism or behavior of other cells possessing functional receptors for the hormone.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
amine biosynthetic process
The chemical reactions and pathways resulting in the formation of any organic compound that is weakly basic in character and contains an amino or a substituted amino group. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
androgen metabolic process
The chemical reactions and pathways involving androgens, C19 steroid hormones that can stimulate the development of male sexual characteristics.
B cell mediated immunity
Any process involved with the carrying out of an immune response by a B cell, through, for instance, the production of antibodies or cytokines, or antigen presentation to T cells.
cellular amino acid biosynthetic process
The chemical reactions and pathways resulting in the formation of amino acids, organic acids containing one or more amino substituents.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
carboxylic acid biosynthetic process
The chemical reactions and pathways resulting in the formation of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
carboxylic acid catabolic process
The chemical reactions and pathways resulting in the breakdown of carboxylic acids, any organic acid containing one or more carboxyl (-COOH) groups.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.
activation of plasma proteins involved in acute inflammatory response
Any process activating plasma proteins by proteolysis as part of an acute inflammatory response.
bile acid metabolic process
The chemical reactions and pathways involving bile acids, any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine.
humoral immune response mediated by circulating immunoglobulin
An immune response dependent upon secreted immunoglobulin. An example of this process is found in Mus musculus.
complement activation
Any process involved in the activation of any of the steps of the complement cascade, which allows for the direct killing of microbes, the disposal of immune complexes, and the regulation of other immune processes; the initial steps of complement activation involve one of three pathways, the classical pathway, the alternative pathway, and the lectin pathway, all of which lead to the terminal complement pathway.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
extracellular region
The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite.
peroxisome
A small, membrane-bounded organelle that uses dioxygen (O2) to oxidize organic molecules; contains some enzymes that produce and others that degrade hydrogen peroxide (H2O2).
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
extracellular space
That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
extracellular region part
Any constituent part of the extracellular region, the space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers constituent parts of the host cell environment outside an intracellular parasite.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
microbody
Cytoplasmic organelles, spherical or oval in shape, that are bounded by a single membrane and contain oxidative enzymes, especially those utilizing hydrogen peroxide (H2O2).

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
metal ion binding
Interacting selectively and non-covalently with any metal ion.
iron ion binding
Interacting selectively and non-covalently with iron (Fe) ions.
amino acid binding
Interacting selectively and non-covalently with an amino acid, organic acids containing one or more amino substituents.
vitamin binding
Interacting selectively and non-covalently with a vitamin, one of a number of unrelated organic substances that occur in many foods in small amounts and that are necessary in trace amounts for the normal metabolic functioning of the body.
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
carboxylic acid binding
Interacting selectively and non-covalently with a carboxylic acid, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
ion binding
Interacting selectively and non-covalently with ions, charged atoms or groups of atoms.
cation binding
Interacting selectively and non-covalently with cations, charged atoms or groups of atoms with a net positive charge.
amine binding
Interacting selectively and non-covalently with any organic compound that is weakly basic in character and contains an amino or a substituted amino group.
transition metal ion binding
Interacting selectively and non-covalently with a transition metal ions; a transition metal is an element whose atom has an incomplete d-subshell of extranuclear electrons, or which gives rise to a cation or cations with an incomplete d-subshell. Transition metals often have more than one valency state. Biologically relevant transition metals include vanadium, manganese, iron, copper, cobalt, nickel, molybdenum and silver.
cofactor binding
Interacting selectively and non-covalently with a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
vitamin B6 binding
Interacting selectively and non-covalently with any of the vitamin B6 compounds: pyridoxal, pyridoxamine and pyridoxine and the active form, pyridoxal phosphate.
all
NA
amino acid binding
Interacting selectively and non-covalently with an amino acid, organic acids containing one or more amino substituents.
pyridoxal phosphate binding
Interacting selectively and non-covalently with pyridoxal 5' phosphate, 3-hydroxy-5-(hydroxymethyl)-2-methyl4-pyridine carboxaldehyde 5' phosphate, the biologically active form of vitamin B6.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04610 | 1.707e-04 | 0.9119 | 8 | 21 | Complement and coagulation cascades |
| 00270 | 2.854e-04 | 0.4777 | 6 | 11 | Cysteine and methionine metabolism |
| 04146 | 1.157e-02 | 1.303 | 7 | 30 | Peroxisome |
AARS2alanyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000124608), score: 0.65 ABCA1ATP-binding cassette, sub-family A (ABC1), member 1 (ENSG00000165029), score: 0.74 ABCA5ATP-binding cassette, sub-family A (ABC1), member 5 (ENSG00000154265), score: -0.68 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.59 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.8 ACOT12acyl-CoA thioesterase 12 (ENSG00000172497), score: 0.72 ACSL5acyl-CoA synthetase long-chain family member 5 (ENSG00000197142), score: 0.66 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.73 ADPGKADP-dependent glucokinase (ENSG00000159322), score: 0.6 AGXTalanine-glyoxylate aminotransferase (ENSG00000172482), score: 0.66 AK2adenylate kinase 2 (ENSG00000004455), score: 0.7 AKR1D1aldo-keto reductase family 1, member D1 (delta 4-3-ketosteroid-5-beta-reductase) (ENSG00000122787), score: 0.75 ALBalbumin (ENSG00000163631), score: 0.66 AMBPalpha-1-microglobulin/bikunin precursor (ENSG00000106927), score: 0.61 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.64 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.68 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: 0.71 ARHGEF38Rho guanine nucleotide exchange factor (GEF) 38 (ENSG00000138784), score: 0.67 ARL14ADP-ribosylation factor-like 14 (ENSG00000179674), score: 0.97 ARL5BADP-ribosylation factor-like 5B (ENSG00000165997), score: 0.7 ASPGasparaginase homolog (S. cerevisiae) (ENSG00000166183), score: 0.59 ATP8B1ATPase, aminophospholipid transporter, class I, type 8B, member 1 (ENSG00000081923), score: 0.61 AVPR1Aarginine vasopressin receptor 1A (ENSG00000166148), score: 0.82 BLMHbleomycin hydrolase (ENSG00000108578), score: 0.59 BMP2bone morphogenetic protein 2 (ENSG00000125845), score: 0.67 C5complement component 5 (ENSG00000106804), score: 0.71 C8Acomplement component 8, alpha polypeptide (ENSG00000157131), score: 0.65 CA6carbonic anhydrase VI (ENSG00000131686), score: 0.75 CBY1chibby homolog 1 (Drosophila) (ENSG00000100211), score: -0.73 CD82CD82 molecule (ENSG00000085117), score: 0.67 CDO1cysteine dioxygenase, type I (ENSG00000129596), score: 0.59 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.68 CLDN1claudin 1 (ENSG00000163347), score: 0.64 CPB2carboxypeptidase B2 (plasma) (ENSG00000080618), score: 0.65 CTBSchitobiase, di-N-acetyl- (ENSG00000117151), score: 0.8 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.62 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.77 CYB5R2cytochrome b5 reductase 2 (ENSG00000166394), score: 0.61 CYP2R1cytochrome P450, family 2, subfamily R, polypeptide 1 (ENSG00000186104), score: 0.59 CYP39A1cytochrome P450, family 39, subfamily A, polypeptide 1 (ENSG00000146233), score: 0.89 DERL1Der1-like domain family, member 1 (ENSG00000136986), score: 0.65 DHTKD1dehydrogenase E1 and transketolase domain containing 1 (ENSG00000181192), score: 0.76 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.68 EGFRepidermal growth factor receptor (ENSG00000146648), score: 0.6 EPHA1EPH receptor A1 (ENSG00000146904), score: 0.64 ESR1estrogen receptor 1 (ENSG00000091831), score: 0.61 F2coagulation factor II (thrombin) (ENSG00000180210), score: 0.62 F9coagulation factor IX (ENSG00000101981), score: 0.74 FETUBfetuin B (ENSG00000090512), score: 0.9 FGF19fibroblast growth factor 19 (ENSG00000162344), score: 0.59 FGGfibrinogen gamma chain (ENSG00000171557), score: 0.62 FN1fibronectin 1 (ENSG00000115414), score: 0.67 GAR1GAR1 ribonucleoprotein homolog (yeast) (ENSG00000109534), score: 0.62 GAS6growth arrest-specific 6 (ENSG00000183087), score: -0.87 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.69 GK5glycerol kinase 5 (putative) (ENSG00000175066), score: 0.74 GNMTglycine N-methyltransferase (ENSG00000124713), score: 0.67 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (ENSG00000120053), score: 0.64 GPLD1glycosylphosphatidylinositol specific phospholipase D1 (ENSG00000112293), score: 0.64 GPX7glutathione peroxidase 7 (ENSG00000116157), score: 0.6 HALhistidine ammonia-lyase (ENSG00000084110), score: 0.86 HAO1hydroxyacid oxidase (glycolate oxidase) 1 (ENSG00000101323), score: 0.61 HEBP1heme binding protein 1 (ENSG00000013583), score: 0.65 HGFhepatocyte growth factor (hepapoietin A; scatter factor) (ENSG00000019991), score: 0.67 HM13histocompatibility (minor) 13 (ENSG00000101294), score: 0.66 HPXhemopexin (ENSG00000110169), score: 0.64 IGFBP1insulin-like growth factor binding protein 1 (ENSG00000146678), score: 0.72 ITGA4integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) (ENSG00000115232), score: 0.7 IYDiodotyrosine deiodinase (ENSG00000009765), score: 0.79 KIAA1432KIAA1432 (ENSG00000107036), score: 0.66 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.99 LEAP2liver expressed antimicrobial peptide 2 (ENSG00000164406), score: 0.6 LECT2leukocyte cell-derived chemotaxin 2 (ENSG00000145826), score: 0.9 LOC100292021similar to thioredoxin peroxidase (ENSG00000123131), score: 0.73 LPIN2lipin 2 (ENSG00000101577), score: 0.6 LRATlecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) (ENSG00000121207), score: 0.66 LRIG3leucine-rich repeats and immunoglobulin-like domains 3 (ENSG00000139263), score: 0.6 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.84 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.61 LY75lymphocyte antigen 75 (ENSG00000054219), score: 0.75 MASP2mannan-binding lectin serine peptidase 2 (ENSG00000009724), score: 0.6 MAT1Amethionine adenosyltransferase I, alpha (ENSG00000151224), score: 0.67 MBNL2muscleblind-like 2 (Drosophila) (ENSG00000139793), score: -0.7 MKLN1muskelin 1, intracellular mediator containing kelch motifs (ENSG00000128585), score: 0.76 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.64 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: 0.6 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.64 MTMR2myotubularin related protein 2 (ENSG00000087053), score: -0.73 MTTPmicrosomal triglyceride transfer protein (ENSG00000138823), score: 0.66 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.65 NCSTNnicastrin (ENSG00000162736), score: 0.59 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: 0.86 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.7 NR5A2nuclear receptor subfamily 5, group A, member 2 (ENSG00000116833), score: 0.62 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.62 NUDT12nudix (nucleoside diphosphate linked moiety X)-type motif 12 (ENSG00000112874), score: 0.6 OIT3oncoprotein induced transcript 3 (ENSG00000138315), score: 0.61 ONECUT1one cut homeobox 1 (ENSG00000169856), score: 0.63 OTCornithine carbamoyltransferase (ENSG00000036473), score: 0.85 P4HA1prolyl 4-hydroxylase, alpha polypeptide I (ENSG00000122884), score: 0.66 P4HTMprolyl 4-hydroxylase, transmembrane (endoplasmic reticulum) (ENSG00000178467), score: -0.75 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.62 PANK1pantothenate kinase 1 (ENSG00000152782), score: 0.6 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.66 PEX11Aperoxisomal biogenesis factor 11 alpha (ENSG00000166821), score: 0.71 PLA2G12Bphospholipase A2, group XIIB (ENSG00000138308), score: 0.64 PM20D1peptidase M20 domain containing 1 (ENSG00000162877), score: 0.6 PPEF2protein phosphatase, EF-hand calcium binding domain 2 (ENSG00000156194), score: 0.74 PROZprotein Z, vitamin K-dependent plasma glycoprotein (ENSG00000126231), score: 0.62 PTPRGprotein tyrosine phosphatase, receptor type, G (ENSG00000144724), score: 0.66 QSOX1quiescin Q6 sulfhydryl oxidase 1 (ENSG00000116260), score: -0.7 RAB33BRAB33B, member RAS oncogene family (ENSG00000172007), score: 0.61 RBP4retinol binding protein 4, plasma (ENSG00000138207), score: 0.62 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.79 RGS2regulator of G-protein signaling 2, 24kDa (ENSG00000116741), score: -0.77 SCARB1scavenger receptor class B, member 1 (ENSG00000073060), score: 0.64 SDC2syndecan 2 (ENSG00000169439), score: 0.62 SERPINA10serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 (ENSG00000140093), score: 0.67 SERPINC1serpin peptidase inhibitor, clade C (antithrombin), member 1 (ENSG00000117601), score: 0.67 SGPL1sphingosine-1-phosphate lyase 1 (ENSG00000166224), score: 0.6 SGSM2small G protein signaling modulator 2 (ENSG00000141258), score: -0.68 SLC16A10solute carrier family 16, member 10 (aromatic amino acid transporter) (ENSG00000112394), score: 0.64 SLC25A17solute carrier family 25 (mitochondrial carrier; peroxisomal membrane protein, 34kDa), member 17 (ENSG00000100372), score: 0.6 SLC25A47solute carrier family 25, member 47 (ENSG00000140107), score: 0.65 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.7 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.66 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.66 SLC38A4solute carrier family 38, member 4 (ENSG00000139209), score: 0.72 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.7 SLC46A1solute carrier family 46 (folate transporter), member 1 (ENSG00000076351), score: 0.74 SLC7A2solute carrier family 7 (cationic amino acid transporter, y+ system), member 2 (ENSG00000003989), score: 0.79 SLC7A6solute carrier family 7 (cationic amino acid transporter, y+ system), member 6 (ENSG00000103064), score: -0.8 SNX24sorting nexin 24 (ENSG00000064652), score: 0.68 SPP2secreted phosphoprotein 2, 24kDa (ENSG00000072080), score: 0.73 SSR3signal sequence receptor, gamma (translocon-associated protein gamma) (ENSG00000114850), score: 0.6 STAB2stabilin 2 (ENSG00000136011), score: 0.6 STARD5StAR-related lipid transfer (START) domain containing 5 (ENSG00000172345), score: 0.81 TATtyrosine aminotransferase (ENSG00000198650), score: 0.72 TBC1D8BTBC1 domain family, member 8B (with GRAM domain) (ENSG00000133138), score: 0.61 TDO2tryptophan 2,3-dioxygenase (ENSG00000151790), score: 0.69 TLR3toll-like receptor 3 (ENSG00000164342), score: 0.63 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.76 TMEM38Atransmembrane protein 38A (ENSG00000072954), score: -0.68 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.68 TOM1L2target of myb1-like 2 (chicken) (ENSG00000175662), score: -0.68 TP53BP1tumor protein p53 binding protein 1 (ENSG00000067369), score: -0.82 TP53INP1tumor protein p53 inducible nuclear protein 1 (ENSG00000164938), score: 0.61 TRAM2translocation associated membrane protein 2 (ENSG00000065308), score: 0.63 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.73 TWISTNBTWIST neighbor (ENSG00000105849), score: 0.75 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: -0.91 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.71 VSIG4V-set and immunoglobulin domain containing 4 (ENSG00000155659), score: 0.7 WDFY2WD repeat and FYVE domain containing 2 (ENSG00000139668), score: 0.66 WDR43WD repeat domain 43 (ENSG00000163811), score: 0.63 WNT9Awingless-type MMTV integration site family, member 9A (ENSG00000143816), score: 0.63 XCR1chemokine (C motif) receptor 1 (ENSG00000173578), score: 0.62 XKR9XK, Kell blood group complex subunit-related family, member 9 (ENSG00000221947), score: 1 ZBTB2zinc finger and BTB domain containing 2 (ENSG00000181472), score: 0.61 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.83 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.6
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |