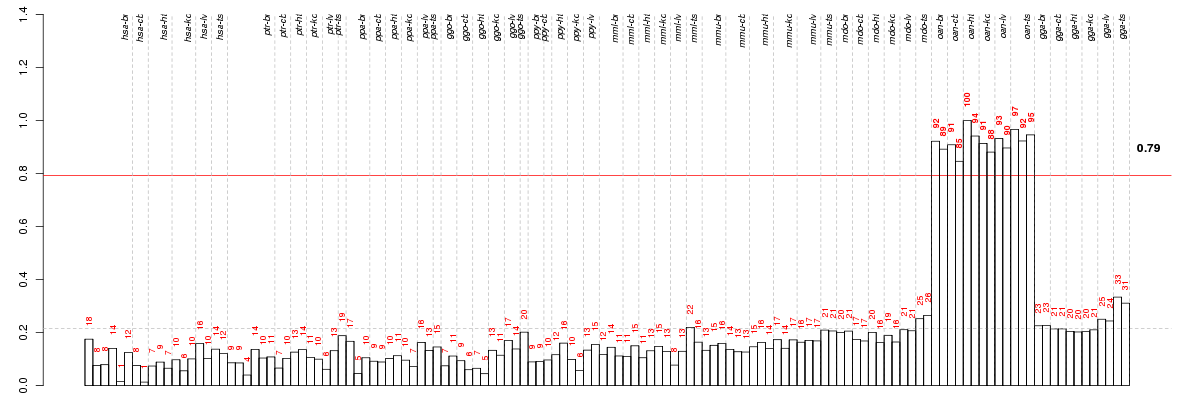

Under-expression is coded with green,
over-expression with red color.

modification-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent modification of the target protein.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
proteolysis
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with the hydrolysis of peptide bonds.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein as part of the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
gene expression
The process by which a gene's sequence is converted into a mature gene product or products (proteins or RNA). This includes the production of an RNA transcript as well as any processing to produce a mature RNA product or an mRNA (for protein-coding genes) and the translation of that mRNA into protein. Some protein processing events may be included when they are required to form an active form of a product from an inactive precursor form.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
modification-dependent macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, initiated by covalent modification of the target molecule.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
nucleic acid metabolic process
Any cellular metabolic process involving nucleic acids.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
cellular macromolecule metabolic process
The chemical reactions and pathways involving macromolecules, any molecule of high relative molecular mass, the structure of which essentially comprises the multiple repetition of units derived, actually or conceptually, from molecules of low relative molecular mass, as carried out by individual cells.
protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general. Includes protein modification.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by the destruction of the native, active configuration, with or without the hydrolysis of peptide bonds.
cellular macromolecule catabolic process
The chemical reactions and pathways resulting in the breakdown of a macromolecule, any large molecule including proteins, nucleic acids and carbohydrates, as carried out by individual cells.
cellular protein metabolic process
The chemical reactions and pathways involving a specific protein, rather than of proteins in general, occurring at the level of an individual cell. Includes protein modification.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
cellular protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
RNA processing
Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules.
RNA metabolic process
The cellular chemical reactions and pathways involving RNA, ribonucleic acid, one of the two main type of nucleic acid, consisting of a long, unbranched macromolecule formed from ribonucleotides joined in 3',5'-phosphodiester linkage.
proteolysis involved in cellular protein catabolic process
The hydrolysis of a peptide bond or bonds within a protein as part of the chemical reactions and pathways resulting in the breakdown of a protein by individual cells.
modification-dependent protein catabolic process
The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent modification of the target protein.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
nucleus
A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent.
nucleoplasm
That part of the nuclear content other than the chromosomes or the nucleolus.
membrane-enclosed lumen
The enclosed volume within a sealed membrane or between two sealed membranes. Encompasses the volume enclosed by the membranes of a particular organelle, e.g. endoplasmic reticulum lumen, or the space between the two lipid bilayers of a double membrane surrounding an organelle, e.g. nuclear envelope lumen.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle lumen
The internal volume enclosed by the membranes of a particular organelle; includes the volume enclosed by a single organelle membrane, e.g. endoplasmic reticulum lumen, or the volume enclosed by the innermost of the two lipid bilayers of an organelle envelope, e.g. nuclear lumen.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
intracellular organelle lumen
An organelle lumen that is part of an intracellular organelle.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
ribonucleoprotein complex
A macromolecular complex containing both protein and RNA molecules.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
nuclear lumen
The volume enclosed by the nuclear inner membrane.
nucleoplasm
That part of the nuclear content other than the chromosomes or the nucleolus.
nuclear part
Any constituent part of the nucleus, a membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
RNA binding
Interacting selectively and non-covalently with an RNA molecule or a portion thereof.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
all
NA
AANATaralkylamine N-acetyltransferase (ENSG00000129673), score: 0.82 AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: 0.65 ACER3alkaline ceramidase 3 (ENSG00000078124), score: 0.66 ACTN4actinin, alpha 4 (ENSG00000130402), score: -0.7 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: -0.6 ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.59 ADALadenosine deaminase-like (ENSG00000168803), score: 0.66 ADSLadenylosuccinate lyase (ENSG00000239900), score: 0.57 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: -0.58 ANAPC2anaphase promoting complex subunit 2 (ENSG00000176248), score: -0.58 ANKRD40ankyrin repeat domain 40 (ENSG00000154945), score: -0.75 AP4E1adaptor-related protein complex 4, epsilon 1 subunit (ENSG00000081014), score: 0.59 AQRaquarius homolog (mouse) (ENSG00000021776), score: 0.78 ARFGAP1ADP-ribosylation factor GTPase activating protein 1 (ENSG00000101199), score: -0.69 ARHGEF18Rho/Rac guanine nucleotide exchange factor (GEF) 18 (ENSG00000104880), score: -0.67 ARHGEF3Rho guanine nucleotide exchange factor (GEF) 3 (ENSG00000163947), score: -0.73 ASF1AASF1 anti-silencing function 1 homolog A (S. cerevisiae) (ENSG00000111875), score: 0.76 ATG13ATG13 autophagy related 13 homolog (S. cerevisiae) (ENSG00000175224), score: 0.59 ATPAF2ATP synthase mitochondrial F1 complex assembly factor 2 (ENSG00000171953), score: 0.62 AZI15-azacytidine induced 1 (ENSG00000141577), score: -0.76 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: -0.69 BTBD2BTB (POZ) domain containing 2 (ENSG00000133243), score: -0.59 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: 0.59 C14orf102chromosome 14 open reading frame 102 (ENSG00000119720), score: -0.73 C14orf126chromosome 14 open reading frame 126 (ENSG00000129480), score: 0.7 C17orf68chromosome 17 open reading frame 68 (ENSG00000178971), score: -0.59 C17orf71chromosome 17 open reading frame 71 (ENSG00000167447), score: 0.64 C1orf25chromosome 1 open reading frame 25 (ENSG00000121486), score: 0.8 C20orf4chromosome 20 open reading frame 4 (ENSG00000131043), score: 0.71 C6orf62chromosome 6 open reading frame 62 (ENSG00000112308), score: 0.81 C6orf89chromosome 6 open reading frame 89 (ENSG00000198663), score: 0.59 C8orf40chromosome 8 open reading frame 40 (ENSG00000176209), score: -0.79 CCDC127coiled-coil domain containing 127 (ENSG00000164366), score: -0.81 CCDC93coiled-coil domain containing 93 (ENSG00000125633), score: 0.73 CDC37L1cell division cycle 37 homolog (S. cerevisiae)-like 1 (ENSG00000106993), score: -0.59 CDC40cell division cycle 40 homolog (S. cerevisiae) (ENSG00000168438), score: 0.6 CDC45cell division cycle 45 homolog (S. cerevisiae) (ENSG00000093009), score: 0.66 CDKN2AIPCDKN2A interacting protein (ENSG00000168564), score: -0.8 CDT1chromatin licensing and DNA replication factor 1 (ENSG00000167513), score: 0.58 CENPHcentromere protein H (ENSG00000153044), score: 0.57 CENPQcentromere protein Q (ENSG00000031691), score: 0.61 CHAC2ChaC, cation transport regulator homolog 2 (E. coli) (ENSG00000143942), score: 0.62 CHAF1Bchromatin assembly factor 1, subunit B (p60) (ENSG00000159259), score: 0.69 CHEK1CHK1 checkpoint homolog (S. pombe) (ENSG00000149554), score: 0.57 CIAO1cytosolic iron-sulfur protein assembly 1 (ENSG00000144021), score: 0.72 CLK4CDC-like kinase 4 (ENSG00000113240), score: -0.74 COG2component of oligomeric golgi complex 2 (ENSG00000135775), score: 0.65 COG7component of oligomeric golgi complex 7 (ENSG00000168434), score: 0.59 COMMD2COMM domain containing 2 (ENSG00000114744), score: 0.6 COPAcoatomer protein complex, subunit alpha (ENSG00000122218), score: -1 COQ2coenzyme Q2 homolog, prenyltransferase (yeast) (ENSG00000173085), score: 0.67 CPLX4complexin 4 (ENSG00000166569), score: 0.78 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.59 CRBNcereblon (ENSG00000113851), score: -0.58 CRKv-crk sarcoma virus CT10 oncogene homolog (avian) (ENSG00000167193), score: -0.64 CXorf23chromosome X open reading frame 23 (ENSG00000173681), score: 0.66 CYHR1cysteine/histidine-rich 1 (ENSG00000187954), score: 0.58 DAZAP1DAZ associated protein 1 (ENSG00000071626), score: -0.64 DCLRE1CDNA cross-link repair 1C (ENSG00000152457), score: 0.64 DCTN4dynactin 4 (p62) (ENSG00000132912), score: 0.78 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: 0.57 DIRC2disrupted in renal carcinoma 2 (ENSG00000138463), score: 0.67 DNTTIP1deoxynucleotidyltransferase, terminal, interacting protein 1 (ENSG00000101457), score: 0.8 DONSONdownstream neighbor of SON (ENSG00000159147), score: 0.61 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.58 DSCC1defective in sister chromatid cohesion 1 homolog (S. cerevisiae) (ENSG00000136982), score: 0.88 E2F4E2F transcription factor 4, p107/p130-binding (ENSG00000205250), score: -0.81 EDC3enhancer of mRNA decapping 3 homolog (S. cerevisiae) (ENSG00000179151), score: 0.61 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.58 ENPP7ectonucleotide pyrophosphatase/phosphodiesterase 7 (ENSG00000182156), score: 0.72 ERCC6Lexcision repair cross-complementing rodent repair deficiency, complementation group 6-like (ENSG00000186871), score: 0.65 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.73 EXO1exonuclease 1 (ENSG00000174371), score: 0.64 EXOSC1exosome component 1 (ENSG00000171311), score: 0.71 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.61 FAM185Afamily with sequence similarity 185, member A (ENSG00000222011), score: -0.63 FAM63Bfamily with sequence similarity 63, member B (ENSG00000128923), score: 0.74 FARP2FERM, RhoGEF and pleckstrin domain protein 2 (ENSG00000006607), score: -0.63 FBXO21F-box protein 21 (ENSG00000135108), score: -0.58 FBXO4F-box protein 4 (ENSG00000151876), score: 0.57 FLNBfilamin B, beta (ENSG00000136068), score: -0.65 FOXP1forkhead box P1 (ENSG00000114861), score: -0.64 FTSJD2FtsJ methyltransferase domain containing 2 (ENSG00000137200), score: -0.6 GABPB1GA binding protein transcription factor, beta subunit 1 (ENSG00000104064), score: 0.6 GOLGA1golgin A1 (ENSG00000136935), score: -0.65 GPN1GPN-loop GTPase 1 (ENSG00000198522), score: 0.8 GPN3GPN-loop GTPase 3 (ENSG00000111231), score: -0.68 GTF2E1general transcription factor IIE, polypeptide 1, alpha 56kDa (ENSG00000153767), score: 0.68 GUCY2Cguanylate cyclase 2C (heat stable enterotoxin receptor) (ENSG00000070019), score: 0.65 HDXhighly divergent homeobox (ENSG00000165259), score: 0.58 HELQhelicase, POLQ-like (ENSG00000163312), score: 0.85 HERPUD2HERPUD family member 2 (ENSG00000122557), score: 0.6 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.58 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.63 HRH4histamine receptor H4 (ENSG00000134489), score: 0.71 HSPB11heat shock protein family B (small), member 11 (ENSG00000081870), score: 0.65 HTThuntingtin (ENSG00000197386), score: -0.73 INTS9integrator complex subunit 9 (ENSG00000104299), score: 0.65 IPO11importin 11 (ENSG00000086200), score: 0.57 KDELC1KDEL (Lys-Asp-Glu-Leu) containing 1 (ENSG00000134901), score: 0.57 KIAA0090KIAA0090 (ENSG00000127463), score: 0.58 KIAA0907KIAA0907 (ENSG00000132680), score: 0.65 KIAA1407KIAA1407 (ENSG00000163617), score: 0.76 KLHL8kelch-like 8 (Drosophila) (ENSG00000145332), score: 0.65 LOC100133692similar to cell division cycle 2-like 1 (PITSLRE proteins) (ENSG00000008128), score: -0.62 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.61 LOC100293905similar to gastrin-releasing peptide receptor (ENSG00000126010), score: 0.71 LOC653155PRP4 pre-mRNA processing factor 4 homolog B (yeast) pseudogene (ENSG00000112739), score: -0.58 LOH12CR1loss of heterozygosity, 12, chromosomal region 1 (ENSG00000165714), score: 0.77 LRRC45leucine rich repeat containing 45 (ENSG00000169683), score: -0.58 MAK16MAK16 homolog (S. cerevisiae) (ENSG00000198042), score: 0.79 MAN1A2mannosidase, alpha, class 1A, member 2 (ENSG00000198162), score: 0.64 MAP2K5mitogen-activated protein kinase kinase 5 (ENSG00000137764), score: -0.65 MCM6minichromosome maintenance complex component 6 (ENSG00000076003), score: 0.65 MED23mediator complex subunit 23 (ENSG00000112282), score: 0.7 MLLT1myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 (ENSG00000130382), score: -0.71 MRPL12mitochondrial ribosomal protein L12 (ENSG00000183093), score: -0.86 MRPL44mitochondrial ribosomal protein L44 (ENSG00000135900), score: 0.58 MRPS30mitochondrial ribosomal protein S30 (ENSG00000112996), score: 0.59 MTA1metastasis associated 1 (ENSG00000182979), score: -0.61 MTAPmethylthioadenosine phosphorylase (ENSG00000099810), score: 0.62 MYNNmyoneurin (ENSG00000085274), score: 0.72 MYO9Bmyosin IXB (ENSG00000099331), score: -0.58 N6AMT2N-6 adenine-specific DNA methyltransferase 2 (putative) (ENSG00000150456), score: 0.68 NMNAT3nicotinamide nucleotide adenylyltransferase 3 (ENSG00000163864), score: -0.58 NPATnuclear protein, ataxia-telangiectasia locus (ENSG00000149308), score: 0.73 NPRL2nitrogen permease regulator-like 2 (S. cerevisiae) (ENSG00000114388), score: -0.7 NT5DC25'-nucleotidase domain containing 2 (ENSG00000168268), score: -0.59 NUFIP1nuclear fragile X mental retardation protein interacting protein 1 (ENSG00000083635), score: 0.57 NUP133nucleoporin 133kDa (ENSG00000069248), score: 0.66 NUP37nucleoporin 37kDa (ENSG00000075188), score: 0.63 ORC3Lorigin recognition complex, subunit 3-like (yeast) (ENSG00000135336), score: 0.63 OSBPL2oxysterol binding protein-like 2 (ENSG00000130703), score: -0.58 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: 0.6 PCGF3polycomb group ring finger 3 (ENSG00000185619), score: -0.64 PDRG1p53 and DNA-damage regulated 1 (ENSG00000088356), score: 0.69 PI15peptidase inhibitor 15 (ENSG00000137558), score: 0.58 PIGXphosphatidylinositol glycan anchor biosynthesis, class X (ENSG00000163964), score: 0.78 PLDNpallidin homolog (mouse) (ENSG00000104164), score: 0.66 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: -0.58 POLGpolymerase (DNA directed), gamma (ENSG00000140521), score: -0.7 POLR3Fpolymerase (RNA) III (DNA directed) polypeptide F, 39 kDa (ENSG00000132664), score: 0.87 PRPF6PRP6 pre-mRNA processing factor 6 homolog (S. cerevisiae) (ENSG00000101161), score: -0.59 PRPH2peripherin 2 (retinal degeneration, slow) (ENSG00000112619), score: 0.63 PSMA3proteasome (prosome, macropain) subunit, alpha type, 3 (ENSG00000100567), score: 0.57 PUS1pseudouridylate synthase 1 (ENSG00000177192), score: -0.64 RAB11ARAB11A, member RAS oncogene family (ENSG00000103769), score: 0.88 RABL3RAB, member of RAS oncogene family-like 3 (ENSG00000144840), score: 0.62 RAD17RAD17 homolog (S. pombe) (ENSG00000152942), score: 0.6 RAD54L2RAD54-like 2 (S. cerevisiae) (ENSG00000164080), score: -0.58 RANBP3RAN binding protein 3 (ENSG00000031823), score: -0.77 RBM45RNA binding motif protein 45 (ENSG00000155636), score: 0.61 RBP2retinol binding protein 2, cellular (ENSG00000114113), score: 0.64 RCHY1ring finger and CHY zinc finger domain containing 1 (ENSG00000163743), score: 0.71 RCOR3REST corepressor 3 (ENSG00000117625), score: -0.61 RECKreversion-inducing-cysteine-rich protein with kazal motifs (ENSG00000122707), score: 0.69 RFC3replication factor C (activator 1) 3, 38kDa (ENSG00000133119), score: 0.63 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: 0.63 RNF185ring finger protein 185 (ENSG00000138942), score: -0.64 RNMTRNA (guanine-7-) methyltransferase (ENSG00000101654), score: 0.75 RNPEParginyl aminopeptidase (aminopeptidase B) (ENSG00000176393), score: -0.78 RPAINRPA interacting protein (ENSG00000129197), score: 0.72 RPS6KC1ribosomal protein S6 kinase, 52kDa, polypeptide 1 (ENSG00000136643), score: 0.61 RRP9ribosomal RNA processing 9, small subunit (SSU) processome component, homolog (yeast) (ENSG00000114767), score: -0.6 SETD5SET domain containing 5 (ENSG00000168137), score: -0.74 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: 0.65 SPASTspastin (ENSG00000021574), score: 0.63 SPATA5L1spermatogenesis associated 5-like 1 (ENSG00000171763), score: 0.76 SPATS2Lspermatogenesis associated, serine-rich 2-like (ENSG00000196141), score: -0.6 SPENspen homolog, transcriptional regulator (Drosophila) (ENSG00000065526), score: -0.75 SPOPLspeckle-type POZ protein-like (ENSG00000144228), score: 0.57 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.71 SRSF5serine/arginine-rich splicing factor 5 (ENSG00000100650), score: -0.86 SS18synovial sarcoma translocation, chromosome 18 (ENSG00000141380), score: 0.63 STOML2stomatin (EPB72)-like 2 (ENSG00000165283), score: -0.94 STOML3stomatin (EPB72)-like 3 (ENSG00000133115), score: 0.63 SUPV3L1suppressor of var1, 3-like 1 (S. cerevisiae) (ENSG00000156502), score: 0.65 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.59 TARSL2threonyl-tRNA synthetase-like 2 (ENSG00000185418), score: -0.58 TBCCtubulin folding cofactor C (ENSG00000124659), score: 0.61 TBCEtubulin folding cofactor E (ENSG00000116957), score: 0.66 TDRD7tudor domain containing 7 (ENSG00000196116), score: -0.78 TIMM10translocase of inner mitochondrial membrane 10 homolog (yeast) (ENSG00000134809), score: 0.66 TMEM110transmembrane protein 110 (ENSG00000213533), score: -0.6 TMEM128transmembrane protein 128 (ENSG00000132406), score: 0.59 TMEM33transmembrane protein 33 (ENSG00000109133), score: 0.64 TMEM60transmembrane protein 60 (ENSG00000135211), score: 0.71 TOM1L1target of myb1 (chicken)-like 1 (ENSG00000141198), score: -0.62 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.67 TPRA1transmembrane protein, adipocyte asscociated 1 (ENSG00000163870), score: -0.65 TRIP13thyroid hormone receptor interactor 13 (ENSG00000071539), score: 0.76 TRNAU1APtRNA selenocysteine 1 associated protein 1 (ENSG00000180098), score: 0.7 TSG101tumor susceptibility gene 101 (ENSG00000074319), score: 0.57 TSNtranslin (ENSG00000211460), score: 0.58 TSTD2thiosulfate sulfurtransferase (rhodanese)-like domain containing 2 (ENSG00000136925), score: -0.72 TTC13tetratricopeptide repeat domain 13 (ENSG00000143643), score: 0.65 TTC27tetratricopeptide repeat domain 27 (ENSG00000018699), score: 0.58 TUBGCP2tubulin, gamma complex associated protein 2 (ENSG00000130640), score: -0.92 UBA6ubiquitin-like modifier activating enzyme 6 (ENSG00000033178), score: 0.73 UBE2CBPubiquitin-conjugating enzyme E2C binding protein (ENSG00000118420), score: 0.62 UBE4Aubiquitination factor E4A (UFD2 homolog, yeast) (ENSG00000110344), score: 0.69 UBL3ubiquitin-like 3 (ENSG00000122042), score: 0.69 UBXN4UBX domain protein 4 (ENSG00000144224), score: 0.64 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: -0.67 UTP15UTP15, U3 small nucleolar ribonucleoprotein, homolog (S. cerevisiae) (ENSG00000164338), score: 0.77 UTP23UTP23, small subunit (SSU) processome component, homolog (yeast) (ENSG00000147679), score: 0.67 VGLL1vestigial like 1 (Drosophila) (ENSG00000102243), score: 0.98 VPS4Bvacuolar protein sorting 4 homolog B (S. cerevisiae) (ENSG00000119541), score: 0.78 WDR26WD repeat domain 26 (ENSG00000162923), score: -0.64 WDR3WD repeat domain 3 (ENSG00000065183), score: 0.85 WDR44WD repeat domain 44 (ENSG00000131725), score: 0.77 WDR6WD repeat domain 6 (ENSG00000178252), score: -0.59 WDR76WD repeat domain 76 (ENSG00000092470), score: 0.79 WNT3Awingless-type MMTV integration site family, member 3A (ENSG00000154342), score: 0.62 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.76 YIPF1Yip1 domain family, member 1 (ENSG00000058799), score: 0.59 YTHDC2YTH domain containing 2 (ENSG00000047188), score: -0.77 ZC3HC1zinc finger, C3HC-type containing 1 (ENSG00000091732), score: 0.59 ZHX1zinc fingers and homeoboxes 1 (ENSG00000165156), score: 0.58 ZMPSTE24zinc metallopeptidase (STE24 homolog, S. cerevisiae) (ENSG00000084073), score: 0.61 ZNF326zinc finger protein 326 (ENSG00000162664), score: 0.72 ZNHIT6zinc finger, HIT type 6 (ENSG00000117174), score: -0.63 ZW10ZW10, kinetochore associated, homolog (Drosophila) (ENSG00000086827), score: 0.7
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_cb_f_ca1 | oan | cb | f | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_br_m_ca1 | oan | br | m | _ |
| oan_ts_m1_ca1 | oan | ts | m | 1 |
| oan_lv_m_ca1 | oan | lv | m | _ |
| oan_ht_f_ca1 | oan | ht | f | _ |
| oan_ts_m2_ca1 | oan | ts | m | 2 |
| oan_ts_m3_ca1 | oan | ts | m | 3 |
| oan_ht_m_ca1 | oan | ht | m | _ |