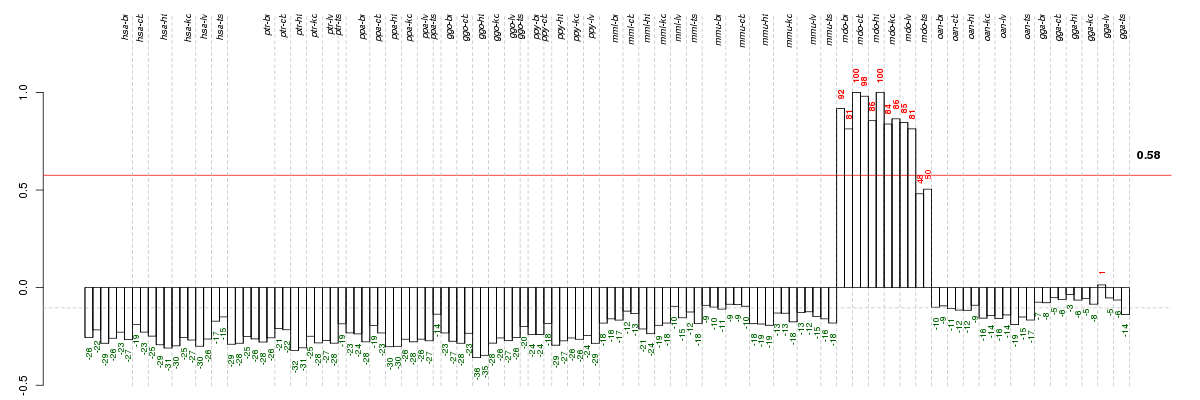

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.

AASDHPPTaminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase (ENSG00000149313), score: -0.62 AATFapoptosis antagonizing transcription factor (ENSG00000108270), score: -0.61 ACANaggrecan (ENSG00000157766), score: 0.52 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: 0.54 ACER2alkaline ceramidase 2 (ENSG00000177076), score: 0.61 ACYP2acylphosphatase 2, muscle type (ENSG00000170634), score: -0.9 ADCK1aarF domain containing kinase 1 (ENSG00000063761), score: -0.74 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: 0.54 ALKBH2alkB, alkylation repair homolog 2 (E. coli) (ENSG00000189046), score: 0.72 AMBRA1autophagy/beclin-1 regulator 1 (ENSG00000110497), score: 0.73 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: -0.61 ANKMY2ankyrin repeat and MYND domain containing 2 (ENSG00000106524), score: 0.55 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.54 ARHGDIGRho GDP dissociation inhibitor (GDI) gamma (ENSG00000167930), score: -0.56 ARSKarylsulfatase family, member K (ENSG00000164291), score: 0.65 ASB1ankyrin repeat and SOCS box-containing 1 (ENSG00000065802), score: 0.69 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.68 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: 0.65 ATP13A3ATPase type 13A3 (ENSG00000133657), score: 0.67 ATP2B4ATPase, Ca++ transporting, plasma membrane 4 (ENSG00000058668), score: -0.84 BCL2L13BCL2-like 13 (apoptosis facilitator) (ENSG00000099968), score: -0.56 BCL7AB-cell CLL/lymphoma 7A (ENSG00000110987), score: 0.5 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.64 BTKBruton agammaglobulinemia tyrosine kinase (ENSG00000010671), score: 0.52 C11orf46chromosome 11 open reading frame 46 (ENSG00000152219), score: 0.61 C11orf57chromosome 11 open reading frame 57 (ENSG00000150776), score: -0.63 C12orf66chromosome 12 open reading frame 66 (ENSG00000174206), score: 0.65 C13orf31chromosome 13 open reading frame 31 (ENSG00000179630), score: 0.81 C14orf101chromosome 14 open reading frame 101 (ENSG00000070269), score: -0.57 C15orf29chromosome 15 open reading frame 29 (ENSG00000134152), score: -0.55 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.51 C17orf75chromosome 17 open reading frame 75 (ENSG00000108666), score: 0.56 C1orf103chromosome 1 open reading frame 103 (ENSG00000121931), score: -0.52 C1orf123chromosome 1 open reading frame 123 (ENSG00000162384), score: 0.7 C20orf43chromosome 20 open reading frame 43 (ENSG00000022277), score: -0.73 C2CD2C2 calcium-dependent domain containing 2 (ENSG00000157617), score: -0.52 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.53 C3orf39chromosome 3 open reading frame 39 (ENSG00000144647), score: 0.69 C3orf59chromosome 3 open reading frame 59 (ENSG00000180611), score: 0.57 C6orf57chromosome 6 open reading frame 57 (ENSG00000154079), score: 0.57 C9orf102chromosome 9 open reading frame 102 (ENSG00000182150), score: -0.59 CAPRIN2caprin family member 2 (ENSG00000110888), score: -0.58 CBFA2T2core-binding factor, runt domain, alpha subunit 2; translocated to, 2 (ENSG00000078699), score: 0.5 CCDC6coiled-coil domain containing 6 (ENSG00000108091), score: 0.51 CCDC77coiled-coil domain containing 77 (ENSG00000120647), score: -0.58 CCDC82coiled-coil domain containing 82 (ENSG00000149231), score: -0.63 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.79 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: -0.57 CDCA3cell division cycle associated 3 (ENSG00000111665), score: 0.82 CDK10cyclin-dependent kinase 10 (ENSG00000185324), score: -0.53 CHD1Lchromodomain helicase DNA binding protein 1-like (ENSG00000131778), score: -0.63 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: -0.65 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.51 CLTBclathrin, light chain B (ENSG00000175416), score: -0.54 COX1cytochrome c oxidase subunit I (ENSG00000198804), score: -0.88 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.56 CSTF1cleavage stimulation factor, 3' pre-RNA, subunit 1, 50kDa (ENSG00000101138), score: -0.51 CWF19L2CWF19-like 2, cell cycle control (S. pombe) (ENSG00000152404), score: -0.54 CYLDcylindromatosis (turban tumor syndrome) (ENSG00000083799), score: -0.64 CYTBcytochrome b (ENSG00000198727), score: -1 CYTH1cytohesin 1 (ENSG00000108669), score: -0.65 DCTDdCMP deaminase (ENSG00000129187), score: -0.52 DCTN5dynactin 5 (p25) (ENSG00000166847), score: -0.52 DCUN1D4DCN1, defective in cullin neddylation 1, domain containing 4 (S. cerevisiae) (ENSG00000109184), score: -0.54 DDX1DEAD (Asp-Glu-Ala-Asp) box polypeptide 1 (ENSG00000079785), score: 0.63 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.61 DENND5BDENN/MADD domain containing 5B (ENSG00000170456), score: 0.49 DERL3Der1-like domain family, member 3 (ENSG00000099958), score: 0.54 DGUOKdeoxyguanosine kinase (ENSG00000114956), score: -0.57 EIF2AK3eukaryotic translation initiation factor 2-alpha kinase 3 (ENSG00000172071), score: 0.5 ERCC6excision repair cross-complementing rodent repair deficiency, complementation group 6 (ENSG00000225830), score: -0.74 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.68 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.62 EXOSC9exosome component 9 (ENSG00000123737), score: 0.57 FAM118Bfamily with sequence similarity 118, member B (ENSG00000197798), score: -0.51 FAM40Bfamily with sequence similarity 40, member B (ENSG00000128578), score: 0.52 FAM73Afamily with sequence similarity 73, member A (ENSG00000180488), score: 0.64 FAM82A2family with sequence similarity 82, member A2 (ENSG00000137824), score: 0.65 FBXO15F-box protein 15 (ENSG00000141665), score: 0.52 FBXO22F-box protein 22 (ENSG00000167196), score: 0.66 FBXO3F-box protein 3 (ENSG00000110429), score: 0.6 FRA10AC1fragile site, folic acid type, rare, fra(10)(q23.3) or fra(10)(q24.2) candidate 1 (ENSG00000148690), score: -0.63 FUT10fucosyltransferase 10 (alpha (1,3) fucosyltransferase) (ENSG00000172728), score: 0.81 GARSglycyl-tRNA synthetase (ENSG00000106105), score: 0.62 GBGT1globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 (ENSG00000148288), score: 0.58 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.61 GLIPR2GLI pathogenesis-related 2 (ENSG00000122694), score: -0.57 GLT8D1glycosyltransferase 8 domain containing 1 (ENSG00000016864), score: -0.57 GMNNgeminin, DNA replication inhibitor (ENSG00000112312), score: -0.52 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.69 GTF3Ageneral transcription factor IIIA (ENSG00000122034), score: -0.62 HAGHhydroxyacylglutathione hydrolase (ENSG00000063854), score: -0.52 HNRNPUheterogeneous nuclear ribonucleoprotein U (scaffold attachment factor A) (ENSG00000153187), score: 0.52 HPRT1hypoxanthine phosphoribosyltransferase 1 (ENSG00000165704), score: 0.5 IKIK cytokine, down-regulator of HLA II (ENSG00000113141), score: -0.75 IKBIPIKBKB interacting protein (ENSG00000166130), score: 0.63 INVSinversin (ENSG00000119509), score: -0.61 IPPintracisternal A particle-promoted polypeptide (ENSG00000197429), score: 0.97 KBTBD8kelch repeat and BTB (POZ) domain containing 8 (ENSG00000163376), score: 0.52 KCTD5potassium channel tetramerisation domain containing 5 (ENSG00000167977), score: -0.66 KIAA0090KIAA0090 (ENSG00000127463), score: 0.5 KIAA0146KIAA0146 (ENSG00000164808), score: -0.54 KIAA0174KIAA0174 (ENSG00000182149), score: -0.6 KIAA1432KIAA1432 (ENSG00000107036), score: 0.5 KLHL5kelch-like 5 (Drosophila) (ENSG00000109790), score: -0.52 LAMP2lysosomal-associated membrane protein 2 (ENSG00000005893), score: -0.55 LEPRE1leucine proline-enriched proteoglycan (leprecan) 1 (ENSG00000117385), score: 0.53 LOC100134387similar to DEAH (Asp-Glu-Ala-His) box polypeptide 40 (ENSG00000108406), score: -0.58 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: 0.57 LYSMD4LysM, putative peptidoglycan-binding, domain containing 4 (ENSG00000183060), score: -0.69 ME2malic enzyme 2, NAD(+)-dependent, mitochondrial (ENSG00000082212), score: -0.69 MED1mediator complex subunit 1 (ENSG00000125686), score: 0.54 MEMO1mediator of cell motility 1 (ENSG00000162961), score: -0.6 MESDC1mesoderm development candidate 1 (ENSG00000140406), score: 0.56 METRNLmeteorin, glial cell differentiation regulator-like (ENSG00000176845), score: -0.62 MID2midline 2 (ENSG00000080561), score: 0.52 MRPL19mitochondrial ribosomal protein L19 (ENSG00000115364), score: 0.67 MRPL33mitochondrial ribosomal protein L33 (ENSG00000158019), score: -0.54 MTIF3mitochondrial translational initiation factor 3 (ENSG00000122033), score: -0.54 N4BP1NEDD4 binding protein 1 (ENSG00000102921), score: 0.68 NAGAN-acetylgalactosaminidase, alpha- (ENSG00000198951), score: 0.59 NARFnuclear prelamin A recognition factor (ENSG00000141562), score: -0.64 NARG2NMDA receptor regulated 2 (ENSG00000128915), score: -0.53 ND2MTND2 (ENSG00000198763), score: -0.96 ND6NADH dehydrogenase, subunit 6 (complex I) (ENSG00000198695), score: -0.58 NFIAnuclear factor I/A (ENSG00000162599), score: 0.53 NIF3L1NIF3 NGG1 interacting factor 3-like 1 (S. pombe) (ENSG00000196290), score: 0.72 NOXO1NADPH oxidase organizer 1 (ENSG00000196408), score: 0.96 NRBF2nuclear receptor binding factor 2 (ENSG00000148572), score: 0.52 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: -0.74 NUP85nucleoporin 85kDa (ENSG00000125450), score: 0.52 OCIAD1OCIA domain containing 1 (ENSG00000109180), score: -0.56 OTOL1otolin 1 homolog (zebrafish) (ENSG00000182447), score: 0.71 PCID2PCI domain containing 2 (ENSG00000126226), score: -0.59 PDE12phosphodiesterase 12 (ENSG00000174840), score: 0.51 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.65 PEX7peroxisomal biogenesis factor 7 (ENSG00000112357), score: 0.61 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.6 PGRprogesterone receptor (ENSG00000082175), score: 0.66 PHF12PHD finger protein 12 (ENSG00000109118), score: 0.64 PHF21APHD finger protein 21A (ENSG00000135365), score: 0.65 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.53 PIGKphosphatidylinositol glycan anchor biosynthesis, class K (ENSG00000142892), score: 0.74 PIGLphosphatidylinositol glycan anchor biosynthesis, class L (ENSG00000108474), score: 0.74 PIGSphosphatidylinositol glycan anchor biosynthesis, class S (ENSG00000087111), score: 0.77 PNPLA6patatin-like phospholipase domain containing 6 (ENSG00000032444), score: 0.49 PODNpodocan (ENSG00000174348), score: 0.63 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.55 PRKACBprotein kinase, cAMP-dependent, catalytic, beta (ENSG00000142875), score: -0.53 PTRH2peptidyl-tRNA hydrolase 2 (ENSG00000141378), score: -0.6 PYCRLpyrroline-5-carboxylate reductase-like (ENSG00000104524), score: 0.64 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.53 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: 0.52 RBM6RNA binding motif protein 6 (ENSG00000004534), score: -0.53 RBPJLrecombination signal binding protein for immunoglobulin kappa J region-like (ENSG00000124232), score: 0.64 RCAN1regulator of calcineurin 1 (ENSG00000159200), score: -0.53 RCAN3RCAN family member 3 (ENSG00000117602), score: 0.5 RG9MTD1RNA (guanine-9-) methyltransferase domain containing 1 (ENSG00000174173), score: -0.6 RIC8Bresistance to inhibitors of cholinesterase 8 homolog B (C. elegans) (ENSG00000111785), score: 0.84 RPS6KA6ribosomal protein S6 kinase, 90kDa, polypeptide 6 (ENSG00000072133), score: 0.69 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.55 SAP30LSAP30-like (ENSG00000164576), score: 0.71 SEC22CSEC22 vesicle trafficking protein homolog C (S. cerevisiae) (ENSG00000093183), score: -0.57 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.57 SFRS8splicing factor, arginine/serine-rich 8 (suppressor-of-white-apricot homolog, Drosophila) (ENSG00000061936), score: 0.72 SIN3BSIN3 homolog B, transcription regulator (yeast) (ENSG00000127511), score: -0.52 SLC15A4solute carrier family 15, member 4 (ENSG00000139370), score: 0.55 SLC35B3solute carrier family 35, member B3 (ENSG00000124786), score: -0.65 SLC38A1solute carrier family 38, member 1 (ENSG00000111371), score: -0.53 SLC38A8solute carrier family 38, member 8 (ENSG00000166558), score: 0.96 SMCR7LSmith-Magenis syndrome chromosome region, candidate 7-like (ENSG00000100335), score: 0.51 SNX13sorting nexin 13 (ENSG00000071189), score: 0.63 SNX8sorting nexin 8 (ENSG00000106266), score: -0.56 SPG20spastic paraplegia 20 (Troyer syndrome) (ENSG00000133104), score: -0.65 SRPX2sushi-repeat-containing protein, X-linked 2 (ENSG00000102359), score: 0.55 STAMBPSTAM binding protein (ENSG00000124356), score: 0.66 STAU2staufen, RNA binding protein, homolog 2 (Drosophila) (ENSG00000040341), score: -0.56 STT3BSTT3, subunit of the oligosaccharyltransferase complex, homolog B (S. cerevisiae) (ENSG00000163527), score: 0.64 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.54 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.5 THYN1thymocyte nuclear protein 1 (ENSG00000151500), score: -0.74 TIAL1TIA1 cytotoxic granule-associated RNA binding protein-like 1 (ENSG00000151923), score: 0.59 TMCO6transmembrane and coiled-coil domains 6 (ENSG00000113119), score: 0.65 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.52 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.71 TMEM184Ctransmembrane protein 184C (ENSG00000164168), score: 0.88 TOE1target of EGR1, member 1 (nuclear) (ENSG00000132773), score: 0.67 TRAF1TNF receptor-associated factor 1 (ENSG00000056558), score: -0.53 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.55 TSNtranslin (ENSG00000211460), score: -0.59 TSPAN13tetraspanin 13 (ENSG00000106537), score: 0.49 TTC12tetratricopeptide repeat domain 12 (ENSG00000149292), score: -0.52 TUBGCP5tubulin, gamma complex associated protein 5 (ENSG00000153575), score: -0.58 TUSC3tumor suppressor candidate 3 (ENSG00000104723), score: 0.5 UBE2G1ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) (ENSG00000132388), score: -0.96 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.61 UCK1uridine-cytidine kinase 1 (ENSG00000130717), score: 0.51 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.69 USP19ubiquitin specific peptidase 19 (ENSG00000172046), score: 0.64 UTP20UTP20, small subunit (SSU) processome component, homolog (yeast) (ENSG00000120800), score: 0.6 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.53 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.49 ZC3H15zinc finger CCCH-type containing 15 (ENSG00000065548), score: -1 ZFYVE27zinc finger, FYVE domain containing 27 (ENSG00000155256), score: 0.53 ZNF346zinc finger protein 346 (ENSG00000113761), score: -0.6 ZNF512zinc finger protein 512 (ENSG00000243943), score: 0.73 ZWILCHZwilch, kinetochore associated, homolog (Drosophila) (ENSG00000174442), score: 0.7
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mdo_br_f_ca1 | mdo | br | f | _ |
| mdo_lv_f_ca1 | mdo | lv | f | _ |
| mdo_kd_m_ca1 | mdo | kd | m | _ |
| mdo_lv_m_ca1 | mdo | lv | m | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| mdo_kd_f_ca1 | mdo | kd | f | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |