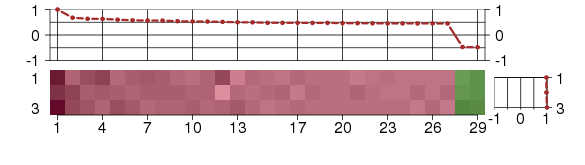

Under-expression is coded with green,
over-expression with red color.



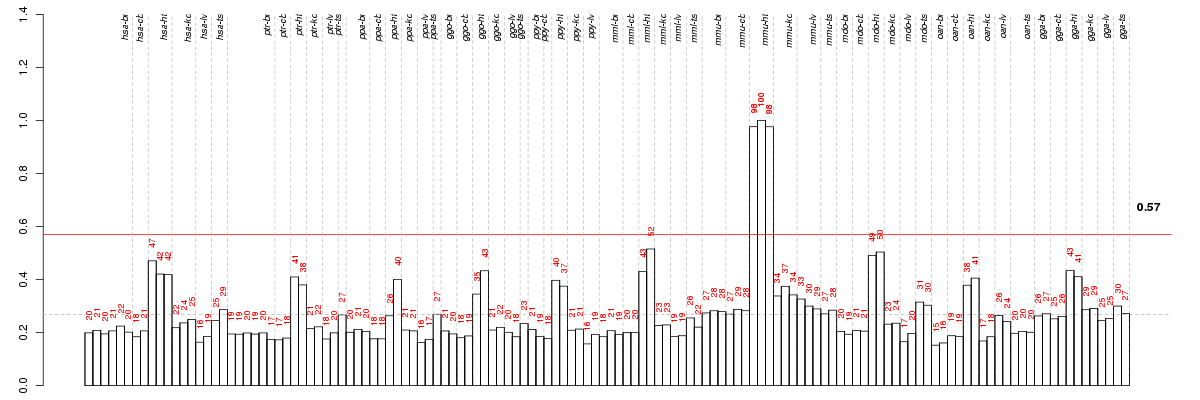
ASB11ankyrin repeat and SOCS box-containing 11 (ENSG00000165192), score: 0.53 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.5 CACNA1Scalcium channel, voltage-dependent, L type, alpha 1S subunit (ENSG00000081248), score: 0.6 CADM1cell adhesion molecule 1 (ENSG00000182985), score: -0.47 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.46 CHRNA10cholinergic receptor, nicotinic, alpha 10 (ENSG00000129749), score: 0.45 CYTL1cytokine-like 1 (ENSG00000170891), score: 0.63 EGLN1egl nine homolog 1 (C. elegans) (ENSG00000135766), score: 0.46 EHD4EH-domain containing 4 (ENSG00000103966), score: 0.45 FAM160A1family with sequence similarity 160, member A1 (ENSG00000164142), score: 0.45 FAM174Bfamily with sequence similarity 174, member B (ENSG00000185442), score: 0.53 H1FXH1 histone family, member X (ENSG00000184897), score: -0.48 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.48 KCNJ5potassium inwardly-rectifying channel, subfamily J, member 5 (ENSG00000120457), score: 0.48 KCNV2potassium channel, subfamily V, member 2 (ENSG00000168263), score: 0.68 LMOD3leiomodin 3 (fetal) (ENSG00000163380), score: 0.56 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.45 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.48 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 1 MFN1mitofusin 1 (ENSG00000171109), score: 0.46 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.57 MYLK4myosin light chain kinase family, member 4 (ENSG00000145949), score: 0.64 MYOCDmyocardin (ENSG00000141052), score: 0.45 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (ENSG00000105953), score: 0.47 PHTF2putative homeodomain transcription factor 2 (ENSG00000006576), score: 0.58 RAPSNreceptor-associated protein of the synapse (ENSG00000165917), score: 0.54 TXLNBtaxilin beta (ENSG00000164440), score: 0.47 UCP3uncoupling protein 3 (mitochondrial, proton carrier) (ENSG00000175564), score: 0.51 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |