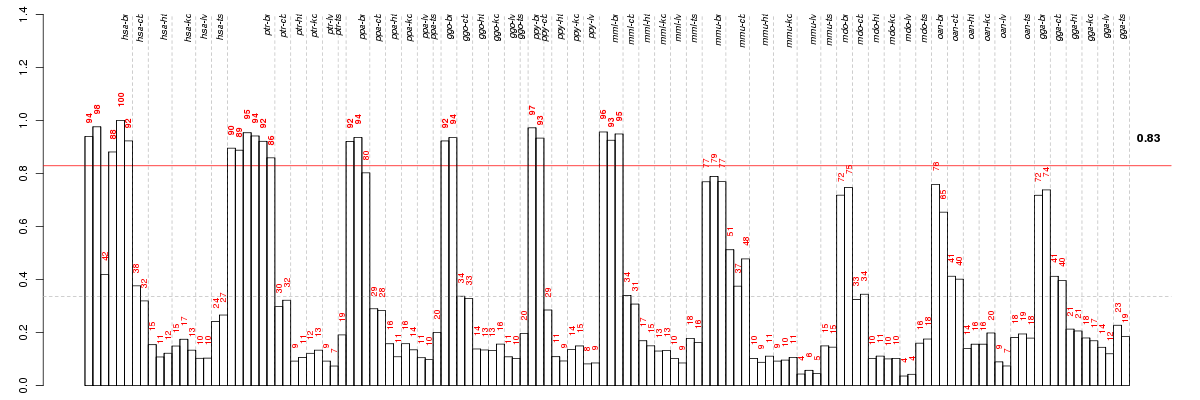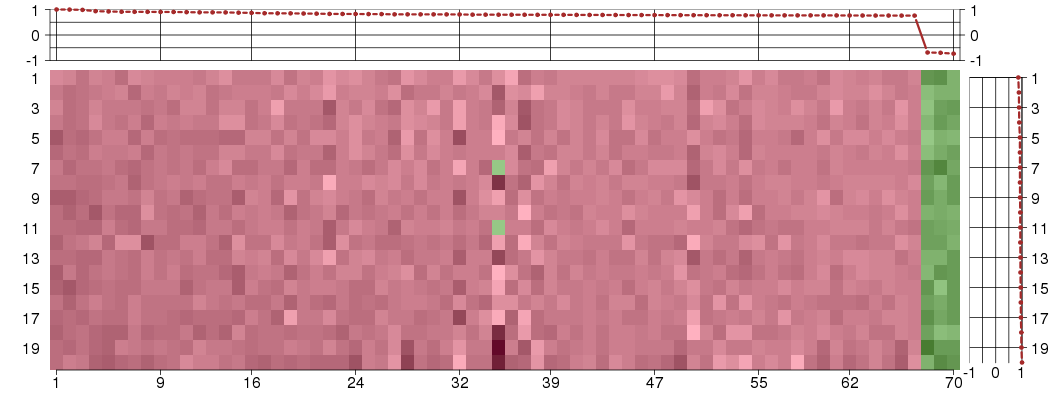

Under-expression is coded with green,
over-expression with red color.

multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
nervous system development
The process whose specific outcome is the progression of nervous tissue over time, from its formation to its mature state.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
all
NA
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
integral to membrane
Penetrating at least one phospholipid bilayer of a membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer. When used to describe a protein, indicates that all or part of the peptide sequence is embedded in the membrane.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
cation channel complex
An ion channel complex through which cations pass.
potassium channel complex
An ion channel complex through which potassium ions pass.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
synapse
The junction between a nerve fiber of one neuron and another neuron or muscle fiber or glial cell; the site of interneuronal communication. As the nerve fiber approaches the synapse it enlarges into a specialized structure, the presynaptic nerve ending, which contains mitochondria and synaptic vesicles. At the tip of the nerve ending is the presynaptic membrane; facing it, and separated from it by a minute cleft (the synaptic cleft) is a specialized area of membrane on the receiving cell, known as the postsynaptic membrane. In response to the arrival of nerve impulses, the presynaptic nerve ending secretes molecules of neurotransmitters into the synaptic cleft. These diffuse across the cleft and transmit the signal to the postsynaptic membrane.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intrinsic to plasma membrane
Located in the plasma membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
ion channel complex
A protein complex that spans a membrane and forms a water-filled channel across the phospholipid bilayer allowing selective ion transport down its electrochemical gradient.
integral to plasma membrane
Penetrating at least one phospholipid bilayer of a plasma membrane. May also refer to the state of being buried in the bilayer with no exposure outside the bilayer.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.
voltage-gated potassium channel complex
A protein complex that forms a transmembrane channel through which potassium ions may cross a cell membrane in response to changes in membrane potential.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
signal transducer activity
Mediates the transfer of a signal from the outside to the inside of a cell by means other than the introduction of the signal molecule itself into the cell.
receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity.
transmembrane receptor activity
Combining with an extracellular or intracellular messenger to initiate a change in cell activity, and spanning to the membrane of either the cell or an organelle.
G-protein coupled receptor activity
A receptor that binds an extracellular ligand and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
transporter activity
Enables the directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
transmembrane transporter activity
Enables the transfer of a substance from one side of a membrane to the other.
G-protein coupled amine receptor activity
A receptor that binds an extracellular amine and transmits the signal to a heterotrimeric G-protein complex. These receptors are characteristically seven-transmembrane receptors and are made up of hetero- or homodimers.
cation transmembrane transporter activity
Catalysis of the transfer of cation from one side of the membrane to the other.
ion transmembrane transporter activity
Catalysis of the transfer of an ion from one side of a membrane to the other.
channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
passive transmembrane transporter activity
Catalysis of the transfer of a solute from one side of the membrane to the other, down the solute's concentration gradient.
voltage-gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel whose open state is dependent on the voltage across the membrane in which it is embedded.
gated channel activity
Catalysis of the transmembrane transfer of a solute by a channel that opens in response to a specific stimulus.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific transporter activity
Enables the directed movement of a specific substance or group of related substances (such as macromolecules, small molecules, ions) into, out of, within or between cells.
molecular transducer activity
The molecular function that accepts an input of one form and creates an output of a different form.
all
NA
substrate-specific transmembrane transporter activity
Enables the transfer of a specific substance or group of related substances from one side of a membrane to the other.
substrate-specific channel activity
Catalysis of energy-independent facilitated diffusion, mediated by passage of a specific solute through a transmembrane aqueous pore or channel. Stereospecificity is not exhibited but this transport may be specific for a particular molecular species or class of molecules.
ion channel activity
Catalysis of facilitated diffusion of an ion (by an energy-independent process) by passage through a transmembrane aqueous pore or channel without evidence for a carrier-mediated mechanism.
cation channel activity
Catalysis of the energy-independent passage of cations across a lipid bilayer down a concentration gradient.
voltage-gated ion channel activity
Catalysis of the transmembrane transfer of an ion by a voltage-gated channel. An ion is an atom or group of atoms carrying an electric charge by virtue of having gained or lost one or more electrons.
voltage-gated cation channel activity
Catalysis of the transmembrane transfer of a cation by a voltage-gated channel. A cation is a positively charged ion.
ADCY2adenylate cyclase 2 (brain) (ENSG00000078295), score: 0.77 C13orf36chromosome 13 open reading frame 36 (ENSG00000180440), score: 0.8 C8orf46chromosome 8 open reading frame 46 (ENSG00000169085), score: 0.89 CACNG3calcium channel, voltage-dependent, gamma subunit 3 (ENSG00000006116), score: 0.76 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.8 CAMK1Gcalcium/calmodulin-dependent protein kinase IG (ENSG00000008118), score: 0.82 CAMK2Acalcium/calmodulin-dependent protein kinase II alpha (ENSG00000070808), score: 0.8 CBLN4cerebellin 4 precursor (ENSG00000054803), score: 0.93 CDH12cadherin 12, type 2 (N-cadherin 2) (ENSG00000154162), score: 0.79 CDH9cadherin 9, type 2 (T1-cadherin) (ENSG00000113100), score: 0.77 CHN1chimerin (chimaerin) 1 (ENSG00000128656), score: 0.79 CHRM3cholinergic receptor, muscarinic 3 (ENSG00000133019), score: 0.85 DLX1distal-less homeobox 1 (ENSG00000144355), score: 0.91 ELFN2extracellular leucine-rich repeat and fibronectin type III domain containing 2 (ENSG00000166897), score: 0.78 ENC1ectodermal-neural cortex 1 (with BTB-like domain) (ENSG00000171617), score: 0.9 ENTPD3ectonucleoside triphosphate diphosphohydrolase 3 (ENSG00000168032), score: 0.77 EPHB6EPH receptor B6 (ENSG00000106123), score: 0.87 FAM19A1family with sequence similarity 19 (chemokine (C-C motif)-like), member A1 (ENSG00000183662), score: 0.89 FAM5Bfamily with sequence similarity 5, member B (ENSG00000198797), score: 0.81 FBLN7fibulin 7 (ENSG00000144152), score: 0.85 FEZF2FEZ family zinc finger 2 (ENSG00000153266), score: 0.91 FOXG1forkhead box G1 (ENSG00000176165), score: 0.85 GLDNgliomedin (ENSG00000186417), score: 0.79 GLRA3glycine receptor, alpha 3 (ENSG00000145451), score: 0.81 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.81 GRIN2Aglutamate receptor, ionotropic, N-methyl D-aspartate 2A (ENSG00000183454), score: 0.77 HRH3histamine receptor H3 (ENSG00000101180), score: 0.76 HTR2A5-hydroxytryptamine (serotonin) receptor 2A (ENSG00000102468), score: 1 KCNA6potassium voltage-gated channel, shaker-related subfamily, member 6 (ENSG00000151079), score: 0.78 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.92 KCNQ5potassium voltage-gated channel, KQT-like subfamily, member 5 (ENSG00000185760), score: 0.99 KCNS1potassium voltage-gated channel, delayed-rectifier, subfamily S, member 1 (ENSG00000124134), score: 1 KCNS2potassium voltage-gated channel, delayed-rectifier, subfamily S, member 2 (ENSG00000156486), score: 0.82 KIAA0020KIAA0020 (ENSG00000080608), score: -0.7 LIMK1LIM domain kinase 1 (ENSG00000106683), score: 0.77 LMO4LIM domain only 4 (ENSG00000143013), score: 0.9 LPPR3lipid phosphate phosphatase-related protein type 3 (ENSG00000129951), score: 0.78 LRFN2leucine rich repeat and fibronectin type III domain containing 2 (ENSG00000156564), score: 0.82 MATKmegakaryocyte-associated tyrosine kinase (ENSG00000007264), score: 0.84 MEF2Cmyocyte enhancer factor 2C (ENSG00000081189), score: 0.79 MGAT5Bmannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase, isozyme B (ENSG00000167889), score: 0.79 MKL2MKL/myocardin-like 2 (ENSG00000186260), score: 0.81 NECAB1N-terminal EF-hand calcium binding protein 1 (ENSG00000123119), score: 0.77 NETO1neuropilin (NRP) and tolloid (TLL)-like 1 (ENSG00000166342), score: 0.89 OCA2oculocutaneous albinism II (ENSG00000104044), score: 0.83 PCSK1proprotein convertase subtilisin/kexin type 1 (ENSG00000175426), score: 0.9 PDE8Bphosphodiesterase 8B (ENSG00000113231), score: 0.84 PLCH1phospholipase C, eta 1 (ENSG00000114805), score: 0.81 PMP2peripheral myelin protein 2 (ENSG00000147588), score: 0.78 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: 0.8 PSDpleckstrin and Sec7 domain containing (ENSG00000059915), score: 0.76 RIMS3regulating synaptic membrane exocytosis 3 (ENSG00000117016), score: 0.79 RORBRAR-related orphan receptor B (ENSG00000198963), score: 0.88 RSPO2R-spondin 2 homolog (Xenopus laevis) (ENSG00000147655), score: 0.91 RXFP1relaxin/insulin-like family peptide receptor 1 (ENSG00000171509), score: 0.77 SATB2SATB homeobox 2 (ENSG00000119042), score: 0.77 SCN3Bsodium channel, voltage-gated, type III, beta (ENSG00000166257), score: 0.78 SERPINI1serpin peptidase inhibitor, clade I (neuroserpin), member 1 (ENSG00000163536), score: 0.78 SNTG1syntrophin, gamma 1 (ENSG00000147481), score: 0.78 SSTsomatostatin (ENSG00000157005), score: 0.78 ST6GALNAC5ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 (ENSG00000117069), score: 0.83 SYT17synaptotagmin XVII (ENSG00000103528), score: 0.79 TOXthymocyte selection-associated high mobility group box (ENSG00000198846), score: 0.76 UNC5Aunc-5 homolog A (C. elegans) (ENSG00000113763), score: 0.77 VSTM2AV-set and transmembrane domain containing 2A (ENSG00000170419), score: 0.77 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.73 ZDHHC22zinc finger, DHHC-type containing 22 (ENSG00000177108), score: 0.81 ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.69 ZMAT4zinc finger, matrin type 4 (ENSG00000165061), score: 0.77 ZNF365zinc finger protein 365 (ENSG00000138311), score: 0.76
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_br_f_ca1 | ptr | br | f | _ |
| hsa_br_m1_ca1 | hsa | br | m | 1 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| ptr_br_m3_ca1 | ptr | br | m | 3 |
| ppa_br_m_ca1 | ppa | br | m | _ |
| ptr_br_m5_ca1 | ptr | br | m | 5 |
| ggo_br_m_ca1 | ggo | br | m | _ |
| hsa_br_f_ca1 | hsa | br | f | _ |
| mml_br_m1_ca1 | mml | br | m | 1 |
| ppy_br_f_ca1 | ppy | br | f | _ |
| ggo_br_f_ca1 | ggo | br | f | _ |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| hsa_br_m2_ca1 | hsa | br | m | 2 |
| ptr_br_m4_ca1 | ptr | br | m | 4 |
| mml_br_f_ca1 | mml | br | f | _ |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| mml_br_m2_ca1 | mml | br | m | 2 |
| ppy_br_m_ca1 | ppy | br | m | _ |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |