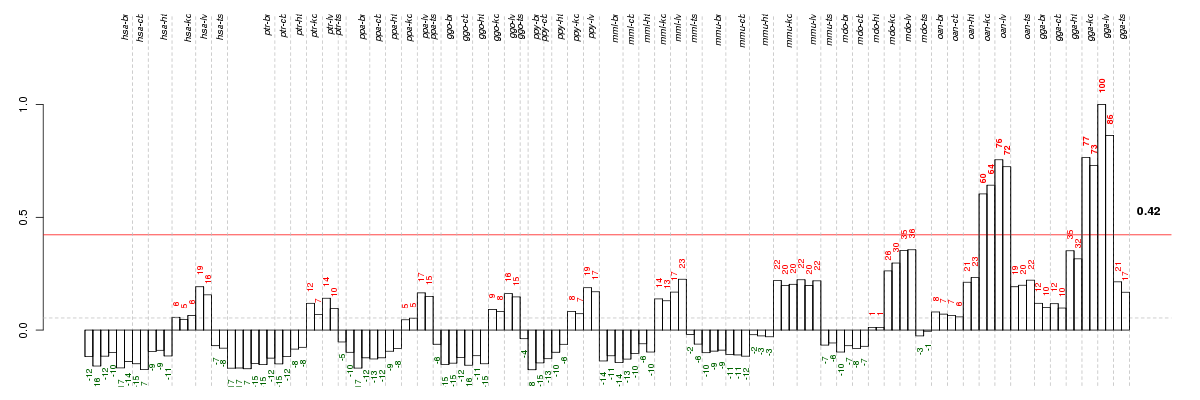

Under-expression is coded with green,
over-expression with red color.

sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
sulfur metabolic process
The chemical reactions and pathways involving the nonmetallic element sulfur or compounds that contain sulfur, such as the amino acids methionine and cysteine or the tripeptide glutathione.
metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
serine family amino acid metabolic process
The chemical reactions and pathways involving amino acids of the serine family, comprising cysteine, glycine, homoserine, selenocysteine and serine.
amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
carboxylic acid metabolic process
The chemical reactions and pathways involving carboxylic acids, any organic acid containing one or more carboxyl (COOH) groups or anions (COO-).
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
organic acid metabolic process
The chemical reactions and pathways involving organic acids, any acidic compound containing carbon in covalent linkage.
cellular amino acid and derivative metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents, and compounds derived from amino acids, as carried out by individual cells.
cellular ketone metabolic process
The chemical reactions and pathways involving any of a class of organic compounds that contain the carbonyl group, CO, and in which the carbonyl group is bonded only to carbon atoms, as carried out by individual cells. The general formula for a ketone is RCOR, where R and R are alkyl or aryl groups.
cellular amine metabolic process
The chemical reactions and pathways involving any organic compound that is weakly basic in character and contains an amino or a substituted amino group, as carried out by individual cells. Amines are called primary, secondary, or tertiary according to whether one, two, or three carbon atoms are attached to the nitrogen atom.
oxoacid metabolic process
The chemical reactions and pathways involving any oxoacid; an oxoacid is a compound which contains oxygen, at least one other element, and at least one hydrogen bound to oxygen, and which produces a conjugate base by loss of positive hydrogen ion(s) (hydrons).
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
sulfur amino acid metabolic process
The chemical reactions and pathways involving amino acids containing sulfur, comprising cysteine, homocysteine, methionine and selenocysteine.
cellular amino acid metabolic process
The chemical reactions and pathways involving amino acids, organic acids containing one or more amino substituents.
cysteine metabolic process
The chemical reactions and pathways involving cysteine, 2-amino-3-mercaptopropanoic acid.


ACVR2Aactivin A receptor, type IIA (ENSG00000121989), score: 0.71 ADSLadenylosuccinate lyase (ENSG00000239900), score: 0.65 AKAP12A kinase (PRKA) anchor protein 12 (ENSG00000131016), score: -0.63 ALDH9A1aldehyde dehydrogenase 9 family, member A1 (ENSG00000143149), score: 0.73 AMDHD1amidohydrolase domain containing 1 (ENSG00000139344), score: 0.6 AP1S3adaptor-related protein complex 1, sigma 3 subunit (ENSG00000152056), score: 0.72 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: 0.61 APBB3amyloid beta (A4) precursor protein-binding, family B, member 3 (ENSG00000113108), score: -0.66 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.58 ASAP1ArfGAP with SH3 domain, ankyrin repeat and PH domain 1 (ENSG00000153317), score: -0.57 C1orf116chromosome 1 open reading frame 116 (ENSG00000182795), score: 0.57 C2orf54chromosome 2 open reading frame 54 (ENSG00000172478), score: 0.57 CA6carbonic anhydrase VI (ENSG00000131686), score: 1 CALML4calmodulin-like 4 (ENSG00000129007), score: 0.54 CAPN2calpain 2, (m/II) large subunit (ENSG00000162909), score: -0.62 CCBL1cysteine conjugate-beta lyase, cytoplasmic (ENSG00000171097), score: 0.58 CD28CD28 molecule (ENSG00000178562), score: 0.57 CD40LGCD40 ligand (ENSG00000102245), score: 0.66 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.63 CDH1cadherin 1, type 1, E-cadherin (epithelial) (ENSG00000039068), score: 0.57 CDO1cysteine dioxygenase, type I (ENSG00000129596), score: 0.56 CEPT1choline/ethanolamine phosphotransferase 1 (ENSG00000134255), score: 0.62 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.64 CLCN4chloride channel 4 (ENSG00000073464), score: -0.58 CLDN1claudin 1 (ENSG00000163347), score: 0.6 CLPXClpX caseinolytic peptidase X homolog (E. coli) (ENSG00000166855), score: 0.62 CRLS1cardiolipin synthase 1 (ENSG00000088766), score: 0.55 CTHcystathionase (cystathionine gamma-lyase) (ENSG00000116761), score: 0.56 CUEDC2CUE domain containing 2 (ENSG00000107874), score: -0.7 CXXC5CXXC finger 5 (ENSG00000171604), score: -0.58 CYB5R2cytochrome b5 reductase 2 (ENSG00000166394), score: 0.56 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.73 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: -0.57 DENND1BDENN/MADD domain containing 1B (ENSG00000213047), score: 0.57 DENND5ADENN/MADD domain containing 5A (ENSG00000184014), score: -0.73 DRAM2DNA-damage regulated autophagy modulator 2 (ENSG00000156171), score: 0.61 EDEM3ER degradation enhancer, mannosidase alpha-like 3 (ENSG00000116406), score: 0.65 EPS15L1epidermal growth factor receptor pathway substrate 15-like 1 (ENSG00000127527), score: -0.6 EREGepiregulin (ENSG00000124882), score: 1 ERLEC1endoplasmic reticulum lectin 1 (ENSG00000068912), score: 0.59 FAF2Fas associated factor family member 2 (ENSG00000113194), score: 0.55 FBXL20F-box and leucine-rich repeat protein 20 (ENSG00000108306), score: 0.64 GCLMglutamate-cysteine ligase, modifier subunit (ENSG00000023909), score: 0.56 HNF4Ghepatocyte nuclear factor 4, gamma (ENSG00000164749), score: 0.74 HPGDShematopoietic prostaglandin D synthase (ENSG00000163106), score: 0.83 ICOSinducible T-cell co-stimulator (ENSG00000163600), score: 0.55 IKZF3IKAROS family zinc finger 3 (Aiolos) (ENSG00000161405), score: 0.55 IL12Binterleukin 12B (natural killer cell stimulatory factor 2, cytotoxic lymphocyte maturation factor 2, p40) (ENSG00000113302), score: 0.7 IL22RA2interleukin 22 receptor, alpha 2 (ENSG00000164485), score: 0.73 KLHDC8Bkelch domain containing 8B (ENSG00000185909), score: -0.63 KMOkynurenine 3-monooxygenase (kynurenine 3-hydroxylase) (ENSG00000117009), score: 0.56 LACTBlactamase, beta (ENSG00000103642), score: 0.59 LAPTM4Blysosomal protein transmembrane 4 beta (ENSG00000104341), score: -0.84 LHX8LIM homeobox 8 (ENSG00000162624), score: 0.59 LRP11low density lipoprotein receptor-related protein 11 (ENSG00000120256), score: -0.65 LRRC15leucine rich repeat containing 15 (ENSG00000172061), score: 0.61 LRRC31leucine rich repeat containing 31 (ENSG00000114248), score: 0.71 LRRC58leucine rich repeat containing 58 (ENSG00000163428), score: 0.63 MAPK8IP3mitogen-activated protein kinase 8 interacting protein 3 (ENSG00000138834), score: -0.59 MARVELD2MARVEL domain containing 2 (ENSG00000152939), score: 0.55 MBD3methyl-CpG binding domain protein 3 (ENSG00000071655), score: -0.65 MPDZmultiple PDZ domain protein (ENSG00000107186), score: -0.62 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.55 MTA1metastasis associated 1 (ENSG00000182979), score: -0.74 NRBP1nuclear receptor binding protein 1 (ENSG00000115216), score: -0.79 OSTalphaorganic solute transporter alpha (ENSG00000163959), score: 0.58 PAFAH2platelet-activating factor acetylhydrolase 2, 40kDa (ENSG00000158006), score: 0.6 PARP6poly (ADP-ribose) polymerase family, member 6 (ENSG00000137817), score: -0.61 PCK2phosphoenolpyruvate carboxykinase 2 (mitochondrial) (ENSG00000100889), score: 0.55 PDX1pancreatic and duodenal homeobox 1 (ENSG00000139515), score: 0.59 PHF16PHD finger protein 16 (ENSG00000102221), score: 0.59 PPP2R2Dprotein phosphatase 2, regulatory subunit B, delta (ENSG00000175470), score: -0.59 PPP2R5Dprotein phosphatase 2, regulatory subunit B', delta (ENSG00000112640), score: -0.69 PYROXD2pyridine nucleotide-disulphide oxidoreductase domain 2 (ENSG00000119943), score: 0.58 RASSF6Ras association (RalGDS/AF-6) domain family member 6 (ENSG00000169435), score: 0.57 RGNregucalcin (senescence marker protein-30) (ENSG00000130988), score: 0.56 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.61 RPIAribose 5-phosphate isomerase A (ENSG00000153574), score: 0.62 SASH3SAM and SH3 domain containing 3 (ENSG00000122122), score: 0.64 SDCCAG8serologically defined colon cancer antigen 8 (ENSG00000054282), score: -0.63 SESN1sestrin 1 (ENSG00000080546), score: 0.56 SGCEsarcoglycan, epsilon (ENSG00000127990), score: -0.67 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.61 SLC2A9solute carrier family 2 (facilitated glucose transporter), member 9 (ENSG00000109667), score: 0.56 SLC30A1solute carrier family 30 (zinc transporter), member 1 (ENSG00000170385), score: 0.65 SLC30A7solute carrier family 30 (zinc transporter), member 7 (ENSG00000162695), score: 0.63 SLC39A9solute carrier family 39 (zinc transporter), member 9 (ENSG00000029364), score: 0.62 SPINK4serine peptidase inhibitor, Kazal type 4 (ENSG00000122711), score: 0.73 SRSF4serine/arginine-rich splicing factor 4 (ENSG00000116350), score: -0.71 SSR1signal sequence receptor, alpha (ENSG00000124783), score: 0.55 ST14suppression of tumorigenicity 14 (colon carcinoma) (ENSG00000149418), score: 0.58 STK24serine/threonine kinase 24 (ENSG00000102572), score: -0.62 STOML3stomatin (EPB72)-like 3 (ENSG00000133115), score: 0.62 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.59 TAAR1trace amine associated receptor 1 (ENSG00000146399), score: 0.78 TADA2Btranscriptional adaptor 2B (ENSG00000173011), score: -0.6 TLR3toll-like receptor 3 (ENSG00000164342), score: 0.55 TM7SF4transmembrane 7 superfamily member 4 (ENSG00000164935), score: 0.67 TMCO3transmembrane and coiled-coil domains 3 (ENSG00000150403), score: -0.58 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.59 TMPRSS13transmembrane protease, serine 13 (ENSG00000137747), score: 0.55 TTC1tetratricopeptide repeat domain 1 (ENSG00000113312), score: -0.66 TTPAtocopherol (alpha) transfer protein (ENSG00000137561), score: 0.57 UBA2ubiquitin-like modifier activating enzyme 2 (ENSG00000126261), score: -0.59 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.58 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: 0.57 WDR41WD repeat domain 41 (ENSG00000164253), score: -0.57 WDR8WD repeat domain 8 (ENSG00000116213), score: -0.61 XPNPEP2X-prolyl aminopeptidase (aminopeptidase P) 2, membrane-bound (ENSG00000122121), score: 0.55 XRCC6BP1XRCC6 binding protein 1 (ENSG00000166896), score: 0.56 YIPF5Yip1 domain family, member 5 (ENSG00000145817), score: 0.6 ZBTB8Azinc finger and BTB domain containing 8A (ENSG00000160062), score: 0.54 ZC3H12Dzinc finger CCCH-type containing 12D (ENSG00000178199), score: 0.58 ZNF704zinc finger protein 704 (ENSG00000164684), score: 0.56
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| oan_kd_m_ca1 | oan | kd | m | _ |
| oan_kd_f_ca1 | oan | kd | f | _ |
| oan_lv_f_ca1 | oan | lv | f | _ |
| gga_kd_f_ca1 | gga | kd | f | _ |
| oan_lv_m_ca1 | oan | lv | m | _ |
| gga_kd_m_ca1 | gga | kd | m | _ |
| gga_lv_f_ca1 | gga | lv | f | _ |
| gga_lv_m_ca1 | gga | lv | m | _ |