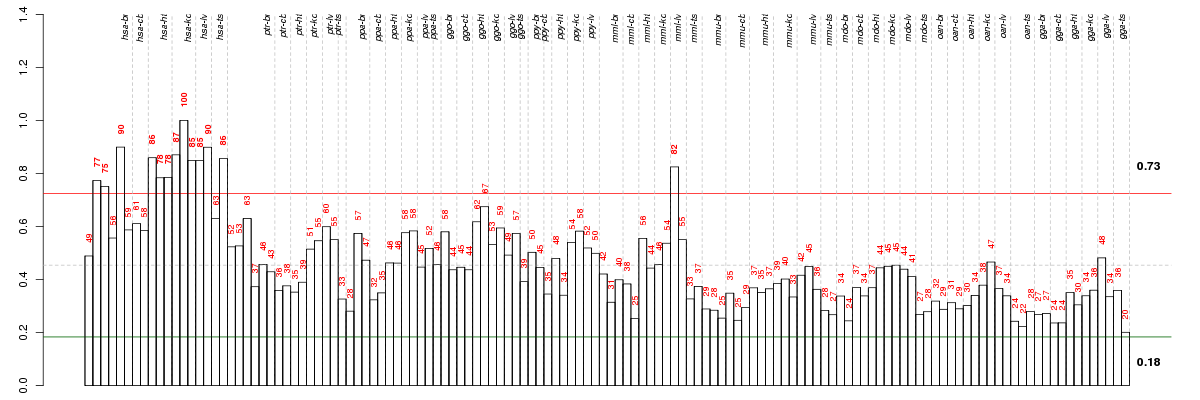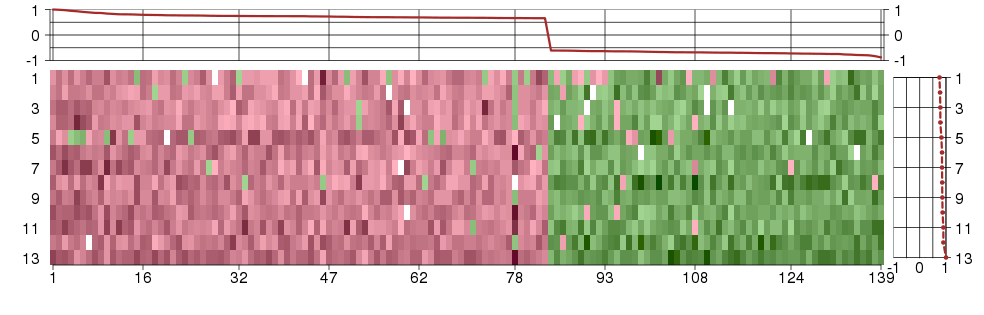

Under-expression is coded with green,
over-expression with red color.


intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.

ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.71 ACAP2ArfGAP with coiled-coil, ankyrin repeat and PH domains 2 (ENSG00000114331), score: -0.71 AIMP2aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 (ENSG00000106305), score: 0.74 ALS2CR4amyotrophic lateral sclerosis 2 (juvenile) chromosome region, candidate 4 (ENSG00000155755), score: -0.69 AP1M1adaptor-related protein complex 1, mu 1 subunit (ENSG00000072958), score: 0.74 APEHN-acylaminoacyl-peptide hydrolase (ENSG00000164062), score: 0.66 APPBP2amyloid beta precursor protein (cytoplasmic tail) binding protein 2 (ENSG00000062725), score: -0.76 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: 0.7 ARID4AAT rich interactive domain 4A (RBP1-like) (ENSG00000032219), score: -0.68 ARRDC1arrestin domain containing 1 (ENSG00000197070), score: 0.67 ATG10ATG10 autophagy related 10 homolog (S. cerevisiae) (ENSG00000152348), score: 0.89 ATG4CATG4 autophagy related 4 homolog C (S. cerevisiae) (ENSG00000125703), score: -0.69 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: 0.67 C19orf56chromosome 19 open reading frame 56 (ENSG00000105583), score: 0.66 C1orf107chromosome 1 open reading frame 107 (ENSG00000117597), score: -0.65 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: 0.67 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.88 CALHM2calcium homeostasis modulator 2 (ENSG00000138172), score: 0.69 CCNT2cyclin T2 (ENSG00000082258), score: -0.67 CD151CD151 molecule (Raph blood group) (ENSG00000177697), score: 0.7 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: 0.81 CHD9chromodomain helicase DNA binding protein 9 (ENSG00000177200), score: -0.72 CIITAclass II, major histocompatibility complex, transactivator (ENSG00000179583), score: 0.99 CISD2CDGSH iron sulfur domain 2 (ENSG00000145354), score: 0.74 CLCN7chloride channel 7 (ENSG00000103249), score: 0.73 CLPTM1LCLPTM1-like (ENSG00000049656), score: 0.74 COMMD2COMM domain containing 2 (ENSG00000114744), score: -0.73 COPEcoatomer protein complex, subunit epsilon (ENSG00000105669), score: 0.75 COQ4coenzyme Q4 homolog (S. cerevisiae) (ENSG00000167113), score: 0.68 CTTNcortactin (ENSG00000085733), score: 0.66 CYB5R3cytochrome b5 reductase 3 (ENSG00000100243), score: 0.74 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (ENSG00000141141), score: -0.66 DLEC1deleted in lung and esophageal cancer 1 (ENSG00000008226), score: 0.68 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.68 DPH1DPH1 homolog (S. cerevisiae) (ENSG00000108963), score: 0.87 EEA1early endosome antigen 1 (ENSG00000102189), score: -0.74 EEFSECeukaryotic elongation factor, selenocysteine-tRNA-specific (ENSG00000132394), score: 0.77 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.77 ENGASEendo-beta-N-acetylglucosaminidase (ENSG00000167280), score: 0.69 ERGIC1endoplasmic reticulum-golgi intermediate compartment (ERGIC) 1 (ENSG00000113719), score: 0.71 EXOSC7exosome component 7 (ENSG00000075914), score: 0.8 FAM135Afamily with sequence similarity 135, member A (ENSG00000082269), score: -0.64 FBXW4F-box and WD repeat domain containing 4 (ENSG00000107829), score: 0.68 FOXP4forkhead box P4 (ENSG00000137166), score: 0.69 GLTPD1glycolipid transfer protein domain containing 1 (ENSG00000224051), score: 0.67 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: 0.77 GPCPD1glycerophosphocholine phosphodiesterase GDE1 homolog (S. cerevisiae) (ENSG00000125772), score: -0.7 GPR114G protein-coupled receptor 114 (ENSG00000159618), score: 0.66 HECTD2HECT domain containing 2 (ENSG00000165338), score: -0.71 HEMK1HemK methyltransferase family member 1 (ENSG00000114735), score: 0.78 HEXDChexosaminidase (glycosyl hydrolase family 20, catalytic domain) containing (ENSG00000169660), score: 0.76 HMBOX1homeobox containing 1 (ENSG00000147421), score: -0.75 HOOK3hook homolog 3 (Drosophila) (ENSG00000168172), score: -0.74 HYAL2hyaluronoglucosaminidase 2 (ENSG00000068001), score: 0.74 INTS2integrator complex subunit 2 (ENSG00000108506), score: -0.7 KRIT1KRIT1, ankyrin repeat containing (ENSG00000001631), score: -0.66 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: -0.72 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: -0.72 LOC652522similar to Werner syndrome protein (ENSG00000165392), score: -0.64 LONP1lon peptidase 1, mitochondrial (ENSG00000196365), score: 0.72 LSM10LSM10, U7 small nuclear RNA associated (ENSG00000181817), score: 0.67 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: 0.7 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (ENSG00000011566), score: -0.77 MAP4K5mitogen-activated protein kinase kinase kinase kinase 5 (ENSG00000012983), score: -0.67 METTL6methyltransferase like 6 (ENSG00000206562), score: -0.83 MMABmethylmalonic aciduria (cobalamin deficiency) cblB type (ENSG00000139428), score: 0.81 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.65 MRPS2mitochondrial ribosomal protein S2 (ENSG00000122140), score: 0.69 MRPS25mitochondrial ribosomal protein S25 (ENSG00000131368), score: 0.75 MVDmevalonate (diphospho) decarboxylase (ENSG00000167508), score: 0.74 NAA25N(alpha)-acetyltransferase 25, NatB auxiliary subunit (ENSG00000111300), score: -0.71 NINJ1ninjurin 1 (ENSG00000131669), score: 0.75 NPLOC4nuclear protein localization 4 homolog (S. cerevisiae) (ENSG00000182446), score: 0.75 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: -0.73 NTHL1nth endonuclease III-like 1 (E. coli) (ENSG00000065057), score: 0.69 ORC6Lorigin recognition complex, subunit 6 like (yeast) (ENSG00000091651), score: -0.64 PABPC1Lpoly(A) binding protein, cytoplasmic 1-like (ENSG00000101104), score: 0.81 PDS5BPDS5, regulator of cohesion maintenance, homolog B (S. cerevisiae) (ENSG00000083642), score: -0.68 PELI1pellino homolog 1 (Drosophila) (ENSG00000197329), score: -0.63 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: 0.79 PFKLphosphofructokinase, liver (ENSG00000141959), score: 0.74 PHF6PHD finger protein 6 (ENSG00000156531), score: -0.62 PIGQphosphatidylinositol glycan anchor biosynthesis, class Q (ENSG00000007541), score: 0.68 PLEKHJ1pleckstrin homology domain containing, family J member 1 (ENSG00000104886), score: 1 POMGNT1protein O-linked mannose beta1,2-N-acetylglucosaminyltransferase (ENSG00000085998), score: 0.68 PSKH1protein serine kinase H1 (ENSG00000159792), score: 0.76 PSMD9proteasome (prosome, macropain) 26S subunit, non-ATPase, 9 (ENSG00000110801), score: 0.74 PTBP2polypyrimidine tract binding protein 2 (ENSG00000117569), score: -0.63 PTGES2prostaglandin E synthase 2 (ENSG00000148334), score: 0.7 QARSglutaminyl-tRNA synthetase (ENSG00000172053), score: 0.78 RB1CC1RB1-inducible coiled-coil 1 (ENSG00000023287), score: -0.69 RER1RER1 retention in endoplasmic reticulum 1 homolog (S. cerevisiae) (ENSG00000157916), score: 0.73 RFXANKregulatory factor X-associated ankyrin-containing protein (ENSG00000064490), score: 0.7 RICTORRPTOR independent companion of MTOR, complex 2 (ENSG00000164327), score: -0.63 RPUSD4RNA pseudouridylate synthase domain containing 4 (ENSG00000165526), score: 0.67 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: -0.68 RSPO1R-spondin homolog (Xenopus laevis) (ENSG00000169218), score: 0.93 SCAMP1secretory carrier membrane protein 1 (ENSG00000085365), score: -0.79 SDF4stromal cell derived factor 4 (ENSG00000078808), score: 0.74 SEC13SEC13 homolog (S. cerevisiae) (ENSG00000157020), score: 0.74 SENP7SUMO1/sentrin specific peptidase 7 (ENSG00000138468), score: -0.61 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.69 SLC18A2solute carrier family 18 (vesicular monoamine), member 2 (ENSG00000165646), score: 0.84 SLC19A1solute carrier family 19 (folate transporter), member 1 (ENSG00000173638), score: 0.77 SLC24A5solute carrier family 24, member 5 (ENSG00000188467), score: -0.67 SLC2A8solute carrier family 2 (facilitated glucose transporter), member 8 (ENSG00000136856), score: 0.7 SLC39A13solute carrier family 39 (zinc transporter), member 13 (ENSG00000165915), score: 0.68 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.64 SNX13sorting nexin 13 (ENSG00000071189), score: -0.8 SNX17sorting nexin 17 (ENSG00000115234), score: 0.73 SNX22sorting nexin 22 (ENSG00000157734), score: 0.95 SPASTspastin (ENSG00000021574), score: -0.63 STARD3StAR-related lipid transfer (START) domain containing 3 (ENSG00000131748), score: 0.66 SUN1Sad1 and UNC84 domain containing 1 (ENSG00000164828), score: 0.68 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: -0.78 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: -0.7 TBC1D8TBC1 domain family, member 8 (with GRAM domain) (ENSG00000204634), score: 0.91 TBL3transducin (beta)-like 3 (ENSG00000183751), score: 0.77 TH1LTH1-like (Drosophila) (ENSG00000101158), score: 0.97 THOC2THO complex 2 (ENSG00000125676), score: -0.63 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.73 TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.75 TMEM79transmembrane protein 79 (ENSG00000163472), score: 0.68 TRDMT1tRNA aspartic acid methyltransferase 1 (ENSG00000107614), score: -0.73 TTC15tetratricopeptide repeat domain 15 (ENSG00000171853), score: 0.86 UNC45Aunc-45 homolog A (C. elegans) (ENSG00000140553), score: 0.71 UPF1UPF1 regulator of nonsense transcripts homolog (yeast) (ENSG00000005007), score: 0.69 URM1ubiquitin related modifier 1 (ENSG00000167118), score: 0.72 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: -0.61 VCPIP1valosin containing protein (p97)/p47 complex interacting protein 1 (ENSG00000175073), score: -0.69 VPS41vacuolar protein sorting 41 homolog (S. cerevisiae) (ENSG00000006715), score: -0.61 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.79 WDR1WD repeat domain 1 (ENSG00000071127), score: 0.75 WDR59WD repeat domain 59 (ENSG00000103091), score: 0.66 YIPF3Yip1 domain family, member 3 (ENSG00000137207), score: 0.83 ZBTB6zinc finger and BTB domain containing 6 (ENSG00000186130), score: -0.75 ZNF292zinc finger protein 292 (ENSG00000188994), score: -0.74 ZNF644zinc finger protein 644 (ENSG00000122482), score: -0.68 ZNF828zinc finger protein 828 (ENSG00000198824), score: -0.61
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m6_ca1 | hsa | br | m | 6 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_ht_m2_ca1 | hsa | ht | m | 2 |
| hsa_ht_f_ca1 | hsa | ht | f | _ |
| mml_lv_m_ca1 | mml | lv | m | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| hsa_lv_m1_ca1 | hsa | lv | m | 1 |
| hsa_ts_m1_ca1 | hsa | ts | m | 1 |
| hsa_ht_m1_ca1 | hsa | ht | m | 1 |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| hsa_lv_m2_ca1 | hsa | lv | m | 2 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |