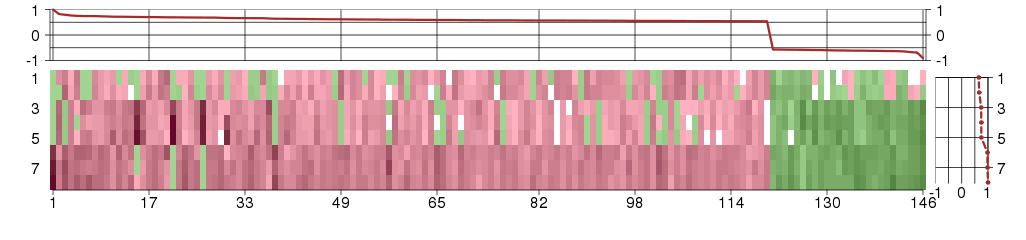

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
acetyl-CoA metabolic process
The chemical reactions and pathways involving acetyl-CoA, a derivative of coenzyme A in which the sulfhydryl group is acetylated; it is a metabolite derived from several pathways (e.g. glycolysis, fatty acid oxidation, amino-acid catabolism) and is further metabolized by the tricarboxylic acid cycle. It is a key intermediate in lipid and terpenoid biosynthesis.
generation of precursor metabolites and energy
The chemical reactions and pathways resulting in the formation of precursor metabolites, substances from which energy is derived, and any process involved in the liberation of energy from these substances.
tricarboxylic acid cycle
A nearly universal metabolic pathway in which the acetyl group of acetyl coenzyme A is effectively oxidized to two CO2 and four pairs of electrons are transferred to coenzymes. The acetyl group combines with oxaloacetate to form citrate, which undergoes successive transformations to isocitrate, 2-oxoglutarate, succinyl-CoA, succinate, fumarate, malate, and oxaloacetate again, thus completing the cycle. In eukaryotes the tricarboxylic acid is confined to the mitochondria. See also glyoxylate cycle.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
coenzyme metabolic process
The chemical reactions and pathways involving coenzymes, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
oxidoreduction coenzyme metabolic process
The chemical reactions and pathways involving coenzymes that are required, in addition to an enzyme and a substrate, for an oxidoreductase reaction to proceed.
NADH metabolic process
The chemical reactions and pathways involving reduced nicotinamide adenine dinucleotide (NADH), a coenzyme present in most living cells and derived from the B vitamin nicotinic acid.
nucleoside phosphate metabolic process
The chemical reactions and pathways involving any phosphorylated nucleoside.
nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds; includes nitrogen fixation, nitrification, denitrification, assimilatory/dissimilatory nitrate reduction and the interconversion of nitrogenous organic matter and ammonium.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, including the breakdown of carbon compounds with the liberation of energy for use by the cell or organism.
aerobic respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which requires oxygen as the terminal electron acceptor.
coenzyme catabolic process
The chemical reactions and pathways resulting in the breakdown of coenzymes, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
nucleotide metabolic process
The chemical reactions and pathways involving a nucleotide, a nucleoside that is esterified with (ortho)phosphate or an oligophosphate at any hydroxyl group on the glycose moiety; may be mono-, di- or triphosphate; this definition includes cyclic nucleotides (nucleoside cyclic phosphates).
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
energy derivation by oxidation of organic compounds
The chemical reactions and pathways by which a cell derives energy from organic compounds; results in the oxidation of the compounds from which energy is released.
pyridine nucleotide metabolic process
The chemical reactions and pathways involving a pyridine nucleotide, a nucleotide characterized by a pyridine derivative as a nitrogen base.
NAD metabolic process
The chemical reactions and pathways involving nicotinamide adenine dinucleotide (NAD), a coenzyme present in most living cells and derived from the B vitamin nicotinic acid.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
primary metabolic process
The chemical reactions and pathways involving those compounds which are formed as a part of the normal anabolic and catabolic processes. These processes take place in most, if not all, cells of the organism.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
small molecule metabolic process
The chemical reactions and pathways involving small molecules, any monomeric molecule of small relative molecular mass.
cellular respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which either requires oxygen (aerobic respiration) or does not (anaerobic respiration).
acetyl-CoA catabolic process
The chemical reactions and pathways resulting in the breakdown of acetyl-CoA, a derivative of coenzyme A in which the sulfhydryl group is acetylated.
nicotinamide nucleotide metabolic process
The chemical reactions and pathways involving nicotinamide nucleotides, any nucleotide that contains combined nicotinamide.
cofactor metabolic process
The chemical reactions and pathways involving a cofactor, a substance that is required for the activity of an enzyme or other protein. Cofactors may be inorganic, such as the metal atoms zinc, iron, and copper in certain forms, or organic, in which case they are referred to as coenzymes. Cofactors may either be bound tightly to active sites or bind loosely with the substrate.
cofactor catabolic process
The chemical reactions and pathways resulting in the breakdown of a cofactor, a substance that is required for the activity of an enzyme or other protein.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
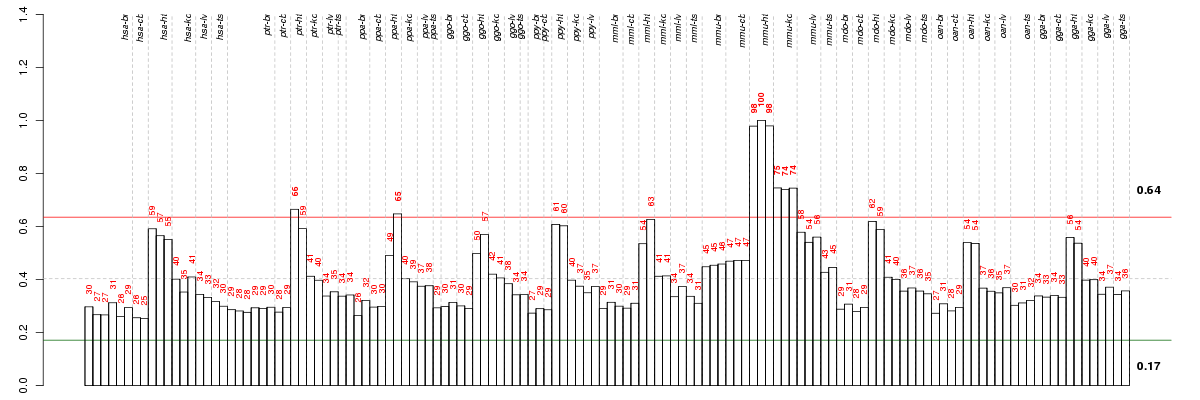
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular nitrogen compound metabolic process
The chemical reactions and pathways involving various organic and inorganic nitrogenous compounds, as carried out by individual cells.
cellular catabolic process
The chemical reactions and pathways resulting in the breakdown of substances, carried out by individual cells.
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
Any cellular metabolic process involving nucleobases, nucleosides, nucleotides and nucleic acids.
cofactor catabolic process
The chemical reactions and pathways resulting in the breakdown of a cofactor, a substance that is required for the activity of an enzyme or other protein.
nucleobase, nucleoside and nucleotide metabolic process
The cellular chemical reactions and pathways involving nucleobases, nucleosides and nucleotides.
coenzyme catabolic process
The chemical reactions and pathways resulting in the breakdown of coenzymes, any of various nonprotein organic cofactors that are required, in addition to an enzyme and a substrate, for an enzymatic reaction to proceed.
acetyl-CoA catabolic process
The chemical reactions and pathways resulting in the breakdown of acetyl-CoA, a derivative of coenzyme A in which the sulfhydryl group is acetylated.
pyridine nucleotide metabolic process
The chemical reactions and pathways involving a pyridine nucleotide, a nucleotide characterized by a pyridine derivative as a nitrogen base.
tricarboxylic acid cycle
A nearly universal metabolic pathway in which the acetyl group of acetyl coenzyme A is effectively oxidized to two CO2 and four pairs of electrons are transferred to coenzymes. The acetyl group combines with oxaloacetate to form citrate, which undergoes successive transformations to isocitrate, 2-oxoglutarate, succinyl-CoA, succinate, fumarate, malate, and oxaloacetate again, thus completing the cycle. In eukaryotes the tricarboxylic acid is confined to the mitochondria. See also glyoxylate cycle.

intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
organelle membrane
The lipid bilayer surrounding an organelle.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
envelope
A multilayered structure surrounding all or part of a cell; encompasses one or more lipid bilayers, and may include a cell wall layer; also includes the space between layers.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.

A4GNTalpha-1,4-N-acetylglucosaminyltransferase (ENSG00000118017), score: 0.69 ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.56 ADAMTS5ADAM metallopeptidase with thrombospondin type 1 motif, 5 (ENSG00000154736), score: 0.55 AFAP1L1actin filament associated protein 1-like 1 (ENSG00000157510), score: 0.57 ANGPT1angiopoietin 1 (ENSG00000154188), score: 0.56 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (ENSG00000148677), score: 0.56 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.61 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.65 ASB11ankyrin repeat and SOCS box-containing 11 (ENSG00000165192), score: 0.75 ASB15ankyrin repeat and SOCS box-containing 15 (ENSG00000146809), score: 0.57 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.72 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.67 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.61 C14orf1chromosome 14 open reading frame 1 (ENSG00000133935), score: -0.61 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.61 CADM1cell adhesion molecule 1 (ENSG00000182985), score: -0.63 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.69 CD274CD274 molecule (ENSG00000120217), score: 0.69 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 0.74 CHPT1choline phosphotransferase 1 (ENSG00000111666), score: 0.57 CLIC5chloride intracellular channel 5 (ENSG00000112782), score: 0.57 CLYBLcitrate lyase beta like (ENSG00000125246), score: 0.54 COL15A1collagen, type XV, alpha 1 (ENSG00000204291), score: 0.54 CWF19L1CWF19-like 1, cell cycle control (S. pombe) (ENSG00000095485), score: -0.57 CYTL1cytokine-like 1 (ENSG00000170891), score: 0.6 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.59 DCP1BDCP1 decapping enzyme homolog B (S. cerevisiae) (ENSG00000151065), score: -0.57 DDOD-aspartate oxidase (ENSG00000203797), score: 0.58 DLDdihydrolipoamide dehydrogenase (ENSG00000091140), score: 0.61 DLSTdihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (ENSG00000119689), score: 0.62 DSPdesmoplakin (ENSG00000096696), score: 0.54 EGLN1egl nine homolog 1 (C. elegans) (ENSG00000135766), score: 0.55 EHD4EH-domain containing 4 (ENSG00000103966), score: 0.61 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.66 ETFDHelectron-transferring-flavoprotein dehydrogenase (ENSG00000171503), score: 0.62 EXOC6exocyst complex component 6 (ENSG00000138190), score: 0.62 FAAHfatty acid amide hydrolase (ENSG00000117480), score: -0.58 FAM160A1family with sequence similarity 160, member A1 (ENSG00000164142), score: 0.63 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: 0.58 FBLIM1filamin binding LIM protein 1 (ENSG00000162458), score: 0.54 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.69 FBXO40F-box protein 40 (ENSG00000163833), score: 0.7 FCHO2FCH domain only 2 (ENSG00000157107), score: 0.55 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.57 FUCA2fucosidase, alpha-L- 2, plasma (ENSG00000001036), score: 0.71 FYCO1FYVE and coiled-coil domain containing 1 (ENSG00000163820), score: 0.57 GALNT1UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 1 (GalNAc-T1) (ENSG00000141429), score: 0.66 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.59 GFM2G elongation factor, mitochondrial 2 (ENSG00000164347), score: 0.57 GOLGA4golgin A4 (ENSG00000144674), score: 0.69 GPX8glutathione peroxidase 8 (putative) (ENSG00000164294), score: 0.54 H1FXH1 histone family, member X (ENSG00000184897), score: -0.91 HBP1HMG-box transcription factor 1 (ENSG00000105856), score: 0.56 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.62 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.68 IARS2isoleucyl-tRNA synthetase 2, mitochondrial (ENSG00000067704), score: 0.6 IDH3Aisocitrate dehydrogenase 3 (NAD+) alpha (ENSG00000166411), score: 0.54 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.64 IL10RBinterleukin 10 receptor, beta (ENSG00000243646), score: 0.61 IL15interleukin 15 (ENSG00000164136), score: 0.6 ITGB6integrin, beta 6 (ENSG00000115221), score: 0.64 IVNS1ABPinfluenza virus NS1A binding protein (ENSG00000116679), score: 0.72 KCNJ5potassium inwardly-rectifying channel, subfamily J, member 5 (ENSG00000120457), score: 0.59 KCNV2potassium channel, subfamily V, member 2 (ENSG00000168263), score: 0.64 KIAA0100KIAA0100 (ENSG00000007202), score: 0.64 KIAA1614KIAA1614 (ENSG00000135835), score: 0.59 KIF13Akinesin family member 13A (ENSG00000137177), score: 0.55 KLHL24kelch-like 24 (Drosophila) (ENSG00000114796), score: 0.7 KRT80keratin 80 (ENSG00000167767), score: 0.65 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: -0.57 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.66 LMOD3leiomodin 3 (fetal) (ENSG00000163380), score: 0.59 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: 0.54 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.56 LRRC14Bleucine rich repeat containing 14B (ENSG00000185028), score: 0.55 LRRC39leucine rich repeat containing 39 (ENSG00000122477), score: 0.55 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 1 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: -0.61 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.56 MAP3K15mitogen-activated protein kinase kinase kinase 15 (ENSG00000180815), score: 0.61 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.6 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.56 MCCC1methylcrotonoyl-CoA carboxylase 1 (alpha) (ENSG00000078070), score: 0.63 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.6 MDH1malate dehydrogenase 1, NAD (soluble) (ENSG00000014641), score: 0.59 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.69 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.58 MEP1Ameprin A, alpha (PABA peptide hydrolase) (ENSG00000112818), score: 0.71 MEP1Bmeprin A, beta (ENSG00000141434), score: 0.68 MFN1mitofusin 1 (ENSG00000171109), score: 0.7 MFN2mitofusin 2 (ENSG00000116688), score: 0.57 MMP13matrix metallopeptidase 13 (collagenase 3) (ENSG00000137745), score: 0.67 MRPL51mitochondrial ribosomal protein L51 (ENSG00000111639), score: 0.56 MRPS23mitochondrial ribosomal protein S23 (ENSG00000181610), score: 0.54 MYL1myosin, light chain 1, alkali; skeletal, fast (ENSG00000168530), score: 0.55 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.55 MYLK4myosin light chain kinase family, member 4 (ENSG00000145949), score: 0.79 MYOCDmyocardin (ENSG00000141052), score: 0.55 NDUFA8NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 8, 19kDa (ENSG00000119421), score: 0.57 NDUFB9NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 9, 22kDa (ENSG00000147684), score: 0.55 NECAB3N-terminal EF-hand calcium binding protein 3 (ENSG00000125967), score: -0.61 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.77 NEK9NIMA (never in mitosis gene a)- related kinase 9 (ENSG00000119638), score: 0.58 NHLRC2NHL repeat containing 2 (ENSG00000196865), score: 0.59 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.67 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.71 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.58 OGDHoxoglutarate (alpha-ketoglutarate) dehydrogenase (lipoamide) (ENSG00000105953), score: 0.73 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.57 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.58 PDHBpyruvate dehydrogenase (lipoamide) beta (ENSG00000168291), score: 0.61 PEMTphosphatidylethanolamine N-methyltransferase (ENSG00000133027), score: -0.6 PHTF2putative homeodomain transcription factor 2 (ENSG00000006576), score: 0.69 PLAAphospholipase A2-activating protein (ENSG00000137055), score: 0.55 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.66 PLXDC2plexin domain containing 2 (ENSG00000120594), score: 0.54 PPP1R3Aprotein phosphatase 1, regulatory (inhibitor) subunit 3A (ENSG00000154415), score: 0.62 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.58 PSMA4proteasome (prosome, macropain) subunit, alpha type, 4 (ENSG00000041357), score: 0.55 PTGR2prostaglandin reductase 2 (ENSG00000140043), score: 0.66 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: 0.58 RALGAPA2Ral GTPase activating protein, alpha subunit 2 (catalytic) (ENSG00000188559), score: 0.57 RAPSNreceptor-associated protein of the synapse (ENSG00000165917), score: 0.66 RBM14RNA binding motif protein 14 (ENSG00000239306), score: -0.59 RNF139ring finger protein 139 (ENSG00000170881), score: 0.62 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.59 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.62 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.63 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.73 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: 0.63 TAB2TGF-beta activated kinase 1/MAP3K7 binding protein 2 (ENSG00000055208), score: 0.57 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.59 TCP11L2t-complex 11 (mouse)-like 2 (ENSG00000166046), score: 0.59 TEAD1TEA domain family member 1 (SV40 transcriptional enhancer factor) (ENSG00000187079), score: 0.74 TIMM17Atranslocase of inner mitochondrial membrane 17 homolog A (yeast) (ENSG00000134375), score: 0.62 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.62 TNFAIP8tumor necrosis factor, alpha-induced protein 8 (ENSG00000145779), score: 0.82 TWSG1twisted gastrulation homolog 1 (Drosophila) (ENSG00000128791), score: 0.54 TXLNBtaxilin beta (ENSG00000164440), score: 0.59 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.68 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.54 UQCRC1ubiquinol-cytochrome c reductase core protein I (ENSG00000010256), score: 0.56 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.56 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.71 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.6 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.74
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ppa_ht_f_ca1 | ppa | ht | f | _ |
| ptr_ht_m_ca1 | ptr | ht | m | _ |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |