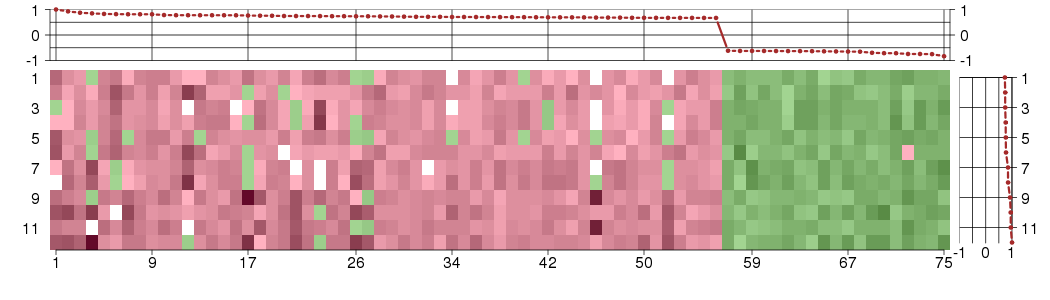

Under-expression is coded with green,
over-expression with red color.


plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
intrinsic to membrane
Located in a membrane such that some covalently attached portion of the gene product, for example part of a peptide sequence or some other covalently attached moiety such as a GPI anchor, spans or is embedded in one or both leaflets of the membrane.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.

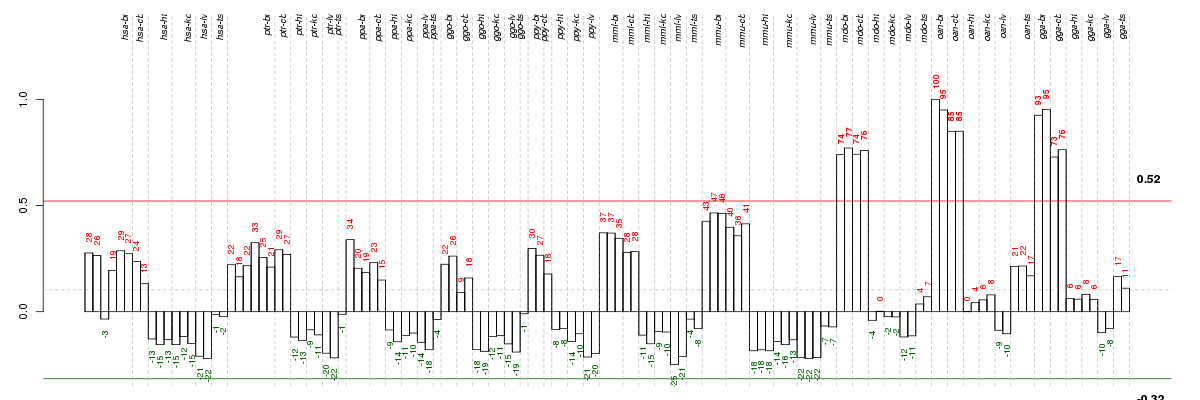
ADCYAP1adenylate cyclase activating polypeptide 1 (pituitary) (ENSG00000141433), score: 0.7 ALG14asparagine-linked glycosylation 14 homolog (S. cerevisiae) (ENSG00000172339), score: -0.71 ARPP21cAMP-regulated phosphoprotein, 21kDa (ENSG00000172995), score: 0.67 AVPR1Barginine vasopressin receptor 1B (ENSG00000198049), score: 0.85 BICD1bicaudal D homolog 1 (Drosophila) (ENSG00000151746), score: 0.75 BMP3bone morphogenetic protein 3 (ENSG00000152785), score: 0.71 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.68 C14orf37chromosome 14 open reading frame 37 (ENSG00000139971), score: 0.68 C15orf60chromosome 15 open reading frame 60 (ENSG00000183324), score: 0.71 C18orf55chromosome 18 open reading frame 55 (ENSG00000075336), score: -0.74 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.7 CASTcalpastatin (ENSG00000153113), score: -0.62 CDK17cyclin-dependent kinase 17 (ENSG00000059758), score: 0.78 CDS2CDP-diacylglycerol synthase (phosphatidate cytidylyltransferase) 2 (ENSG00000101290), score: 0.72 CLCN3chloride channel 3 (ENSG00000109572), score: 0.7 CLPTM1LCLPTM1-like (ENSG00000049656), score: -0.63 CLVS2clavesin 2 (ENSG00000146352), score: 0.7 CNR1cannabinoid receptor 1 (brain) (ENSG00000118432), score: 0.67 CNTN4contactin 4 (ENSG00000144619), score: 0.67 CNTN5contactin 5 (ENSG00000149972), score: 0.93 COL25A1collagen, type XXV, alpha 1 (ENSG00000188517), score: 0.69 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.63 CTNND1catenin (cadherin-associated protein), delta 1 (ENSG00000198561), score: -0.71 CUL4Acullin 4A (ENSG00000139842), score: -0.74 DGKIdiacylglycerol kinase, iota (ENSG00000157680), score: 0.74 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.74 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.76 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.77 EPHX4epoxide hydrolase 4 (ENSG00000172031), score: 0.67 EPS15epidermal growth factor receptor pathway substrate 15 (ENSG00000085832), score: 0.69 FHDC1FH2 domain containing 1 (ENSG00000137460), score: 0.78 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.75 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.74 GAD2glutamate decarboxylase 2 (pancreatic islets and brain, 65kDa) (ENSG00000136750), score: 0.69 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 1 GRB10growth factor receptor-bound protein 10 (ENSG00000106070), score: -0.83 GRID2glutamate receptor, ionotropic, delta 2 (ENSG00000152208), score: 0.7 HHLA1HERV-H LTR-associating 1 (ENSG00000132297), score: 0.73 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.87 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.74 ITFG1integrin alpha FG-GAP repeat containing 1 (ENSG00000129636), score: 0.74 KCNJ6potassium inwardly-rectifying channel, subfamily J, member 6 (ENSG00000157542), score: 0.81 KIAA2022KIAA2022 (ENSG00000050030), score: 0.83 KLHL20kelch-like 20 (Drosophila) (ENSG00000076321), score: 0.73 LEMD2LEM domain containing 2 (ENSG00000161904), score: -0.63 LGI2leucine-rich repeat LGI family, member 2 (ENSG00000153012), score: 0.77 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.74 LOC100293817similar to hCG1811893 (ENSG00000144460), score: 0.67 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.71 LRFN3leucine rich repeat and fibronectin type III domain containing 3 (ENSG00000126243), score: 0.68 LRRC26leucine rich repeat containing 26 (ENSG00000184709), score: 0.73 LRRC33leucine rich repeat containing 33 (ENSG00000174004), score: -0.63 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.66 MAPK8mitogen-activated protein kinase 8 (ENSG00000107643), score: 0.67 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.78 NPY5Rneuropeptide Y receptor Y5 (ENSG00000164129), score: 0.78 NTSneurotensin (ENSG00000133636), score: 0.81 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.76 PRCPprolylcarboxypeptidase (angiotensinase C) (ENSG00000137509), score: -0.63 PTP4A2protein tyrosine phosphatase type IVA, member 2 (ENSG00000184007), score: -0.63 RGS8regulator of G-protein signaling 8 (ENSG00000135824), score: 0.7 RNF114ring finger protein 114 (ENSG00000124226), score: -0.65 SERPINB2serpin peptidase inhibitor, clade B (ovalbumin), member 2 (ENSG00000197632), score: 0.78 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.82 SLC32A1solute carrier family 32 (GABA vesicular transporter), member 1 (ENSG00000101438), score: 0.7 SLC6A9solute carrier family 6 (neurotransmitter transporter, glycine), member 9 (ENSG00000196517), score: 0.73 SNAP23synaptosomal-associated protein, 23kDa (ENSG00000092531), score: -0.63 STX12syntaxin 12 (ENSG00000117758), score: 0.69 TFAP2Dtranscription factor AP-2 delta (activating enhancer binding protein 2 delta) (ENSG00000008197), score: 0.67 TGM2transglutaminase 2 (C polypeptide, protein-glutamine-gamma-glutamyltransferase) (ENSG00000198959), score: -0.64 THSD7Athrombospondin, type I, domain containing 7A (ENSG00000005108), score: 0.81 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: -0.64 TPBGtrophoblast glycoprotein (ENSG00000146242), score: 0.67 TRPC1transient receptor potential cation channel, subfamily C, member 1 (ENSG00000144935), score: 0.71
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_cb_m_ca1 | gga | cb | m | _ |
| mdo_br_m_ca1 | mdo | br | m | _ |
| mdo_cb_m_ca1 | mdo | cb | m | _ |
| mdo_cb_f_ca1 | mdo | cb | f | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |
| mdo_br_f_ca1 | mdo | br | f | _ |
| oan_cb_m_ca1 | oan | cb | m | _ |
| oan_cb_f_ca1 | oan | cb | f | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| oan_br_f_ca1 | oan | br | f | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| oan_br_m_ca1 | oan | br | m | _ |