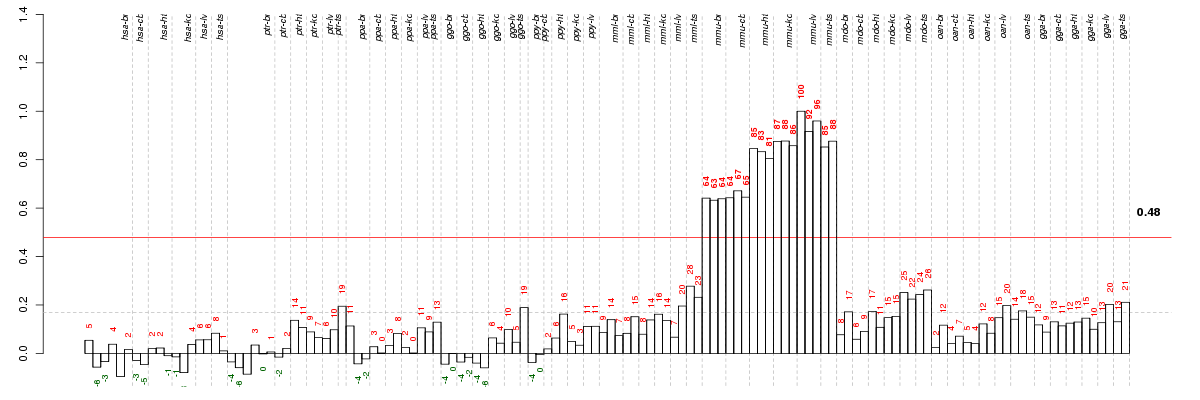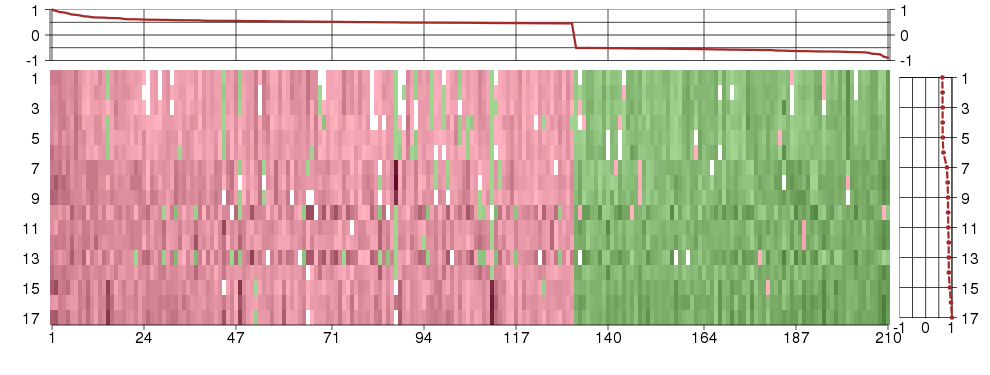

Under-expression is coded with green,
over-expression with red color.



ABCD3ATP-binding cassette, sub-family D (ALD), member 3 (ENSG00000117528), score: 0.47 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: 0.55 AFF4AF4/FMR2 family, member 4 (ENSG00000072364), score: 0.46 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.6 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: 0.53 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.66 ANKRD16ankyrin repeat domain 16 (ENSG00000134461), score: 0.53 ANO5anoctamin 5 (ENSG00000171714), score: -0.54 AP3B1adaptor-related protein complex 3, beta 1 subunit (ENSG00000132842), score: 0.52 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.62 ARFGEF2ADP-ribosylation factor guanine nucleotide-exchange factor 2 (brefeldin A-inhibited) (ENSG00000124198), score: 0.46 ARFIP2ADP-ribosylation factor interacting protein 2 (ENSG00000132254), score: -0.63 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.67 ARSBarylsulfatase B (ENSG00000113273), score: 0.47 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.68 ATG4AATG4 autophagy related 4 homolog A (S. cerevisiae) (ENSG00000101844), score: -0.51 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.56 ATP9BATPase, class II, type 9B (ENSG00000166377), score: 0.49 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.64 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.65 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.56 BPNT13'(2'), 5'-bisphosphate nucleotidase 1 (ENSG00000162813), score: 0.5 BRAPBRCA1 associated protein (ENSG00000089234), score: 0.47 C15orf17chromosome 15 open reading frame 17 (ENSG00000178761), score: -0.53 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.5 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.56 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.46 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.68 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.62 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.63 C4orf34chromosome 4 open reading frame 34 (ENSG00000163683), score: 0.46 C5orf28chromosome 5 open reading frame 28 (ENSG00000151881), score: 0.52 C5orf51chromosome 5 open reading frame 51 (ENSG00000205765), score: 0.46 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.49 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.48 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.73 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 1 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.58 CHMP2Bchromatin modifying protein 2B (ENSG00000083937), score: 0.5 CLEC3BC-type lectin domain family 3, member B (ENSG00000163815), score: -0.53 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.67 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.56 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.48 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: -0.58 CTNNBIP1catenin, beta interacting protein 1 (ENSG00000178585), score: -0.51 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.62 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.74 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.48 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.5 DCTDdCMP deaminase (ENSG00000129187), score: -0.51 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: 0.45 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.66 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.46 DTNBP1dystrobrevin binding protein 1 (ENSG00000047579), score: 0.48 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.69 FABP2fatty acid binding protein 2, intestinal (ENSG00000145384), score: 0.47 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: 0.48 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.73 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: 0.51 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.54 FAM199Xfamily with sequence similarity 199, X-linked (ENSG00000123575), score: -0.65 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: -0.55 FAM76Afamily with sequence similarity 76, member A (ENSG00000009780), score: 0.51 FARS2phenylalanyl-tRNA synthetase 2, mitochondrial (ENSG00000145982), score: 0.49 FBXL7F-box and leucine-rich repeat protein 7 (ENSG00000183580), score: -0.51 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.58 FRS2fibroblast growth factor receptor substrate 2 (ENSG00000166225), score: 0.52 GABARAPL2GABA(A) receptor-associated protein-like 2 (ENSG00000034713), score: -0.54 GADD45Bgrowth arrest and DNA-damage-inducible, beta (ENSG00000099860), score: -0.53 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.54 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.58 GM2AGM2 ganglioside activator (ENSG00000196743), score: 0.48 GNA12guanine nucleotide binding protein (G protein) alpha 12 (ENSG00000146535), score: 0.5 GNA13guanine nucleotide binding protein (G protein), alpha 13 (ENSG00000120063), score: 0.47 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.77 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.58 H1FXH1 histone family, member X (ENSG00000184897), score: -0.85 HDAC8histone deacetylase 8 (ENSG00000147099), score: -0.52 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.68 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.69 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.54 HMBOX1homeobox containing 1 (ENSG00000147421), score: 0.49 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.65 IFFO1intermediate filament family orphan 1 (ENSG00000010295), score: -0.54 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.61 ING1inhibitor of growth family, member 1 (ENSG00000153487), score: 0.49 ITM2Aintegral membrane protein 2A (ENSG00000078596), score: -0.52 KCNRGpotassium channel regulator (ENSG00000198553), score: -0.53 KIAA0100KIAA0100 (ENSG00000007202), score: 0.61 KIAA2018KIAA2018 (ENSG00000176542), score: 0.5 KLF8Kruppel-like factor 8 (ENSG00000102349), score: -0.54 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.55 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: 0.53 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.59 LRSAM1leucine rich repeat and sterile alpha motif containing 1 (ENSG00000148356), score: -0.51 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.5 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.62 MAP3K14mitogen-activated protein kinase kinase kinase 14 (ENSG00000006062), score: -0.54 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.66 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.48 MBIPMAP3K12 binding inhibitory protein 1 (ENSG00000151332), score: -0.57 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.65 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: 0.46 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.62 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.8 METT10Dmethyltransferase 10 domain containing (ENSG00000127804), score: -0.59 MFN1mitofusin 1 (ENSG00000171109), score: 0.54 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.53 MICAL1microtubule associated monoxygenase, calponin and LIM domain containing 1 (ENSG00000135596), score: -0.52 MICALL2MICAL-like 2 (ENSG00000164877), score: -0.51 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.51 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.59 MKS1Meckel syndrome, type 1 (ENSG00000011143), score: -0.53 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.46 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.57 MRPS11mitochondrial ribosomal protein S11 (ENSG00000181991), score: -0.55 MTMR6myotubularin related protein 6 (ENSG00000139505), score: 0.55 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.54 NAA35N(alpha)-acetyltransferase 35, NatC auxiliary subunit (ENSG00000135040), score: 0.56 NCBP1nuclear cap binding protein subunit 1, 80kDa (ENSG00000136937), score: 0.54 ND3NADH dehydrogenase, subunit 3 (complex I) (ENSG00000198840), score: -0.61 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.9 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.89 NSMAFneutral sphingomyelinase (N-SMase) activation associated factor (ENSG00000035681), score: 0.47 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: 0.56 NTHL1nth endonuclease III-like 1 (E. coli) (ENSG00000065057), score: -0.57 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.58 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.52 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.58 OXNAD1oxidoreductase NAD-binding domain containing 1 (ENSG00000154814), score: 0.49 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.63 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.65 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.46 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: -0.64 PGAP3post-GPI attachment to proteins 3 (ENSG00000161395), score: -0.58 PHKA2phosphorylase kinase, alpha 2 (liver) (ENSG00000044446), score: -0.53 PKD2L2polycystic kidney disease 2-like 2 (ENSG00000078795), score: 0.47 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.79 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.5 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: -0.57 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.61 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.59 PPP6R2protein phosphatase 6, regulatory subunit 2 (ENSG00000100239), score: -0.51 PQLC3PQ loop repeat containing 3 (ENSG00000162976), score: -0.56 PROX2prospero homeobox 2 (ENSG00000119608), score: 0.52 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: -0.56 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.6 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: -0.59 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (ENSG00000152229), score: 0.58 PTPLAD2protein tyrosine phosphatase-like A domain containing 2 (ENSG00000188921), score: 0.51 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: 0.57 RDXradixin (ENSG00000137710), score: 0.48 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.66 RFWD3ring finger and WD repeat domain 3 (ENSG00000168411), score: 0.51 RNF139ring finger protein 139 (ENSG00000170881), score: 0.69 RPS19BP1ribosomal protein S19 binding protein 1 (ENSG00000187051), score: -0.59 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.56 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.56 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.63 SART3squamous cell carcinoma antigen recognized by T cells 3 (ENSG00000075856), score: -0.55 SEC23IPSEC23 interacting protein (ENSG00000107651), score: 0.52 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: -0.67 SEP1515 kDa selenoprotein (ENSG00000183291), score: 0.48 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.58 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.51 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.64 SLC25A29solute carrier family 25, member 29 (ENSG00000197119), score: -0.51 SLC30A6solute carrier family 30 (zinc transporter), member 6 (ENSG00000152683), score: 0.47 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.48 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.46 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.76 SNX11sorting nexin 11 (ENSG00000002919), score: -0.53 SNX17sorting nexin 17 (ENSG00000115234), score: -0.53 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.58 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.85 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: 0.55 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.55 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.61 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.58 TGFBRAP1transforming growth factor, beta receptor associated protein 1 (ENSG00000135966), score: 0.47 TM2D2TM2 domain containing 2 (ENSG00000169490), score: 0.49 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.48 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.53 TMEM135transmembrane protein 135 (ENSG00000166575), score: 0.47 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.67 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.73 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.52 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: 0.51 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: 0.46 TUBE1tubulin, epsilon 1 (ENSG00000074935), score: -0.59 TULP4tubby like protein 4 (ENSG00000130338), score: 0.48 TXNDC16thioredoxin domain containing 16 (ENSG00000087301), score: 0.54 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.68 TYW3tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) (ENSG00000162623), score: -0.63 UBE2Wubiquitin-conjugating enzyme E2W (putative) (ENSG00000104343), score: 0.53 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.54 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.58 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.5 USPL1ubiquitin specific peptidase like 1 (ENSG00000132952), score: 0.46 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (ENSG00000197969), score: 0.46 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: 0.52 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: -0.54 WBP4WW domain binding protein 4 (formin binding protein 21) (ENSG00000120688), score: 0.54 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.6 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.89 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: 0.48 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.59 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.94 ZCCHC4zinc finger, CCHC domain containing 4 (ENSG00000168228), score: -0.52 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.55
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mmu_cb_m2_ca1 | mmu | cb | m | 2 |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_ts_m1_ca1 | mmu | ts | m | 1 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_ts_m2_ca1 | mmu | ts | m | 2 |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |