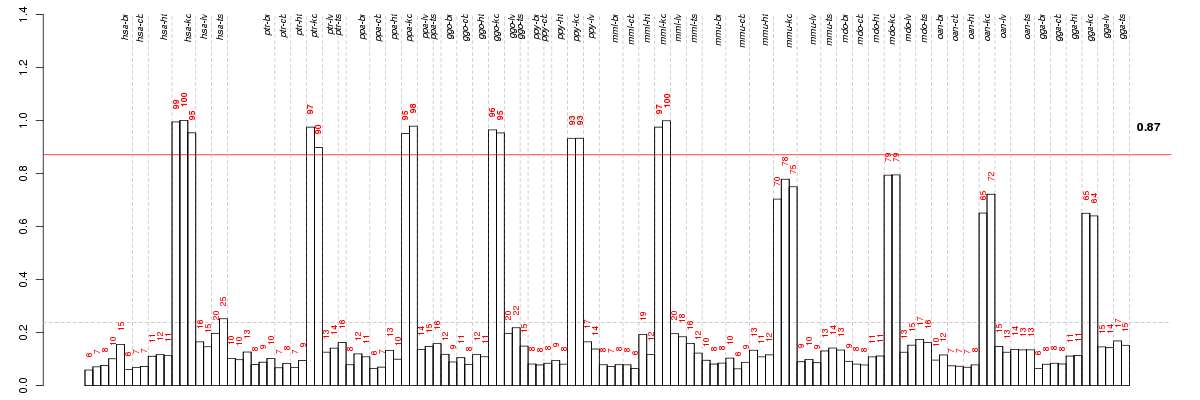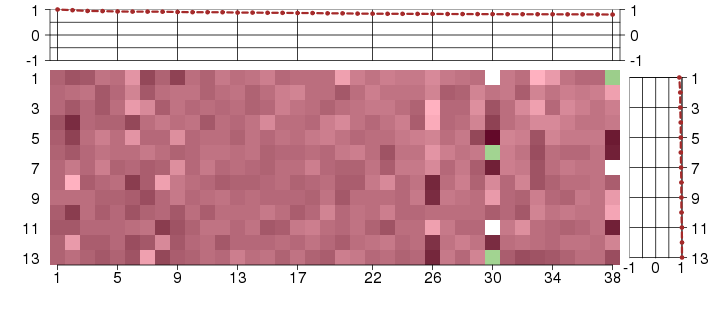

Under-expression is coded with green,
over-expression with red color.

system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
secretion
The controlled release of a substance by a cell, a group of cells, or a tissue.
transport
The directed movement of substances (such as macromolecules, small molecules, ions) into, out of, within or between cells, or within a multicellular organism by means of some external agent such as a transporter or pore.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
homeostatic process
Any biological process involved in the maintenance of an internal steady-state.
ion homeostasis
Any process involved in the maintenance of an internal steady-state of ions within an organism or cell.
chemical homeostasis
Any biological process involved in the maintenance of an internal steady-state of a chemical.
localization
Any process by which a cell, a substance, or a cellular entity, such as a protein complex or organelle, is transported to, and/or maintained in a specific location.
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
cation homeostasis
Any process involved in the maintenance of an internal steady-state of cations within an organism or cell.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
establishment of localization
The directed movement of a cell, substance or cellular entity, such as a protein complex or organelle, to a specific location.
excretion
The elimination by an organism of the waste products that arise as a result of metabolic activity. These products include water, carbon dioxide (CO2), and nitrogenous compounds.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
basolateral plasma membrane
The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
apical part of cell
The region of a polarized cell that forms a tip or is distal to a base. For example, in a polarized epithelial cell, the apical region has an exposed surface and lies opposite to the basal lamina that separates the epithelium from other tissue.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
apical plasma membrane
The region of the plasma membrane located at the apical end of the cell.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
sequence-specific DNA binding transcription factor activity
Interacting selectively and non-covalently with a specific DNA sequence in order to modulate transcription. The transcription factor may or may not also interact selectively with a protein or macromolecular complex.
all
NA
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04960 | 1.636e-02 | 0.1448 | 3 | 16 | Aldosterone-regulated sodium reabsorption |
ACPPacid phosphatase, prostate (ENSG00000014257), score: 0.91 ATP6V0A4ATPase, H+ transporting, lysosomal V0 subunit a4 (ENSG00000105929), score: 0.81 CA12carbonic anhydrase XII (ENSG00000074410), score: 0.89 CLDN16claudin 16 (ENSG00000113946), score: 0.85 DPEP1dipeptidase 1 (renal) (ENSG00000015413), score: 0.85 ELF5E74-like factor 5 (ets domain transcription factor) (ENSG00000135374), score: 0.92 F2RL1coagulation factor II (thrombin) receptor-like 1 (ENSG00000164251), score: 0.88 FOXI1forkhead box I1 (ENSG00000168269), score: 0.94 GATA3GATA binding protein 3 (ENSG00000107485), score: 0.83 GHRHRgrowth hormone releasing hormone receptor (ENSG00000106128), score: 0.82 GRHL2grainyhead-like 2 (Drosophila) (ENSG00000083307), score: 0.81 HOXA10homeobox A10 (ENSG00000153807), score: 0.89 HOXB7homeobox B7 (ENSG00000120087), score: 0.82 HOXD4homeobox D4 (ENSG00000170166), score: 0.88 HSD11B2hydroxysteroid (11-beta) dehydrogenase 2 (ENSG00000176387), score: 0.89 IMPA2inositol(myo)-1(or 4)-monophosphatase 2 (ENSG00000141401), score: 0.82 KCNJ1potassium inwardly-rectifying channel, subfamily J, member 1 (ENSG00000151704), score: 0.92 LMX1BLIM homeobox transcription factor 1, beta (ENSG00000136944), score: 0.9 MMP7matrix metallopeptidase 7 (matrilysin, uterine) (ENSG00000137673), score: 0.97 NOX4NADPH oxidase 4 (ENSG00000086991), score: 0.81 NPHS2nephrosis 2, idiopathic, steroid-resistant (podocin) (ENSG00000116218), score: 1 PAPPA2pappalysin 2 (ENSG00000116183), score: 0.8 PLA2R1phospholipase A2 receptor 1, 180kDa (ENSG00000153246), score: 0.82 PLCG2phospholipase C, gamma 2 (phosphatidylinositol-specific) (ENSG00000197943), score: 0.86 PNPLA1patatin-like phospholipase domain containing 1 (ENSG00000180316), score: 0.83 PROM2prominin 2 (ENSG00000155066), score: 0.94 RHCGRh family, C glycoprotein (ENSG00000140519), score: 0.84 SCNN1Asodium channel, nonvoltage-gated 1 alpha (ENSG00000111319), score: 0.81 SIM2single-minded homolog 2 (Drosophila) (ENSG00000159263), score: 0.81 SLC26A7solute carrier family 26, member 7 (ENSG00000147606), score: 0.92 SLC5A12solute carrier family 5 (sodium/glucose cotransporter), member 12 (ENSG00000148942), score: 0.87 SLC6A19solute carrier family 6 (neutral amino acid transporter), member 19 (ENSG00000174358), score: 0.81 SLC9A4solute carrier family 9 (sodium/hydrogen exchanger), member 4 (ENSG00000180251), score: 0.81 STK32Bserine/threonine kinase 32B (ENSG00000152953), score: 0.86 TFCP2L1transcription factor CP2-like 1 (ENSG00000115112), score: 0.87 TINAGtubulointerstitial nephritis antigen (ENSG00000137251), score: 0.84 TRPV4transient receptor potential cation channel, subfamily V, member 4 (ENSG00000111199), score: 0.83 WDR72WD repeat domain 72 (ENSG00000166415), score: 0.83
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ptr_kd_f_ca1 | ptr | kd | f | _ |
| ppy_kd_m_ca1 | ppy | kd | m | _ |
| ppy_kd_f_ca1 | ppy | kd | f | _ |
| ppa_kd_m_ca1 | ppa | kd | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| hsa_kd_f_ca1 | hsa | kd | f | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| mml_kd_m_ca1 | mml | kd | m | _ |
| ptr_kd_m_ca1 | ptr | kd | m | _ |
| ppa_kd_f_ca1 | ppa | kd | f | _ |
| hsa_kd_m2_ca1 | hsa | kd | m | 2 |
| mml_kd_f_ca1 | mml | kd | f | _ |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |