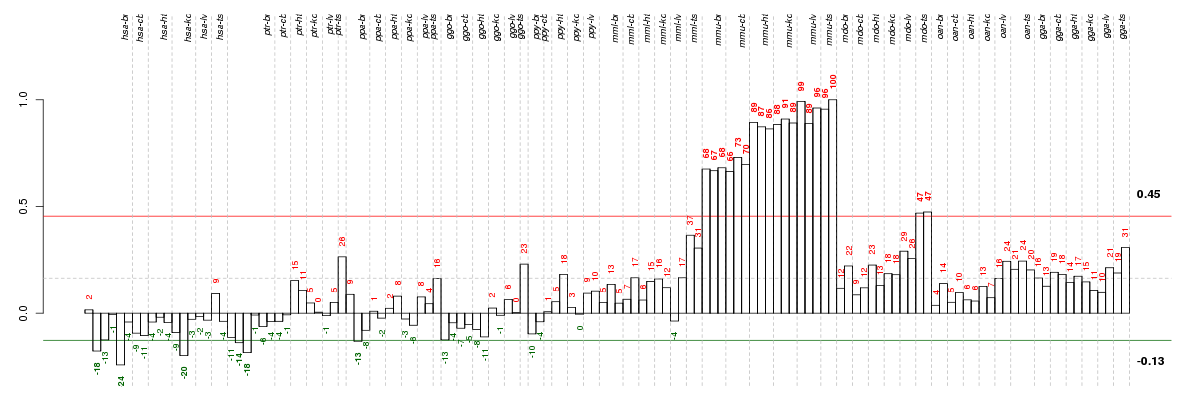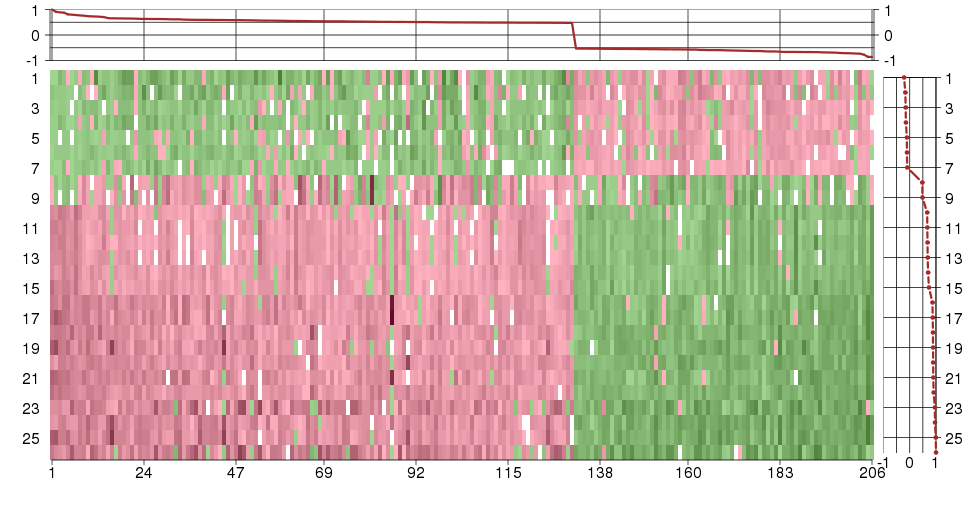

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
ubiquitin-protein ligase activity
Catalysis of the reaction: ATP + ubiquitin + protein lysine = AMP + diphosphate + protein N-ubiquityllysine.
hydrolase activity
Catalysis of the hydrolysis of various bonds, e.g. C-O, C-N, C-C, phosphoric anhydride bonds, etc. Hydrolase is the systematic name for any enzyme of EC class 3.
hydrolase activity, acting on glycosyl bonds
Catalysis of the hydrolysis of any glycosyl bond.
hydrolase activity, hydrolyzing N-glycosyl compounds
Catalysis of the hydrolysis of any N-glycosyl bond.
ligase activity
Catalysis of the ligation of two substances with concomitant breaking of a diphosphate linkage, usually in a nucleoside triphosphate. Ligase is the systematic name for any enzyme of EC class 6.
ligase activity, forming carbon-nitrogen bonds
Catalysis of the ligation of two substances via a carbon-nitrogen bond with concomitant breakage of a diphosphate linkage, usually in a nucleoside triphosphate.
acid-amino acid ligase activity
Catalysis of the ligation of an acid to an amino acid via a carbon-nitrogen bond with concomitant breakage of a diphosphate linkage, usually in a nucleoside triphosphate.
small conjugating protein ligase activity
Catalysis of ATP-dependent isopeptide bond formation between the carboxy-terminal residues of a small conjugating protein such as ubiquitin or a ubiquitin-like protein, and a substrate lysine residue. This function may be performed alone or in conjunction with an E3, ubiquitin-like protein ligase.
all
NA
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: 0.5 ACTR8ARP8 actin-related protein 8 homolog (yeast) (ENSG00000113812), score: 0.54 ADPRHADP-ribosylarginine hydrolase (ENSG00000144843), score: 0.48 AMMECR1LAMME chromosomal region gene 1-like (ENSG00000144233), score: 0.61 ANAPC4anaphase promoting complex subunit 4 (ENSG00000053900), score: 0.56 ANGEL2angel homolog 2 (Drosophila) (ENSG00000174606), score: 0.7 AP3B1adaptor-related protein complex 3, beta 1 subunit (ENSG00000132842), score: 0.55 ARFGEF1ADP-ribosylation factor guanine nucleotide-exchange factor 1(brefeldin A-inhibited) (ENSG00000066777), score: 0.63 ARFIP2ADP-ribosylation factor interacting protein 2 (ENSG00000132254), score: -0.67 ARMC7armadillo repeat containing 7 (ENSG00000125449), score: -0.69 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.67 ATP6V0A2ATPase, H+ transporting, lysosomal V0 subunit a2 (ENSG00000185344), score: 0.59 ATPAF1ATP synthase mitochondrial F1 complex assembly factor 1 (ENSG00000123472), score: -0.66 B4GALT2UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 (ENSG00000117411), score: -0.66 BCL10B-cell CLL/lymphoma 10 (ENSG00000142867), score: 0.58 BPGM2,3-bisphosphoglycerate mutase (ENSG00000172331), score: 0.55 BRAPBRCA1 associated protein (ENSG00000089234), score: 0.51 C10orf118chromosome 10 open reading frame 118 (ENSG00000165813), score: 0.47 C10orf18chromosome 10 open reading frame 18 (ENSG00000108021), score: 0.51 C15orf17chromosome 15 open reading frame 17 (ENSG00000178761), score: -0.54 C17orf39chromosome 17 open reading frame 39 (ENSG00000141034), score: 0.54 C19orf56chromosome 19 open reading frame 56 (ENSG00000105583), score: -0.54 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.6 C1orf58chromosome 1 open reading frame 58 (ENSG00000162819), score: 0.48 C1orf83chromosome 1 open reading frame 83 (ENSG00000116205), score: 0.51 C1orf91chromosome 1 open reading frame 91 (ENSG00000160055), score: 0.64 C21orf63chromosome 21 open reading frame 63 (ENSG00000166979), score: -0.54 C3orf37chromosome 3 open reading frame 37 (ENSG00000183624), score: -0.61 C3orf75chromosome 3 open reading frame 75 (ENSG00000163832), score: -0.67 C5orf28chromosome 5 open reading frame 28 (ENSG00000151881), score: 0.58 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: 0.51 C5orf51chromosome 5 open reading frame 51 (ENSG00000205765), score: 0.48 C7orf23chromosome 7 open reading frame 23 (ENSG00000135185), score: 0.48 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: 0.57 CCDC41coiled-coil domain containing 41 (ENSG00000173588), score: 0.54 CCRL1chemokine (C-C motif) receptor-like 1 (ENSG00000129048), score: 0.74 CDK8cyclin-dependent kinase 8 (ENSG00000132964), score: 1 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: 0.48 CGRRF1cell growth regulator with ring finger domain 1 (ENSG00000100532), score: 0.58 CHMP2Bchromatin modifying protein 2B (ENSG00000083937), score: 0.49 CLEC3BC-type lectin domain family 3, member B (ENSG00000163815), score: -0.59 CNOT6CCR4-NOT transcription complex, subunit 6 (ENSG00000113300), score: 0.49 CNR2cannabinoid receptor 2 (macrophage) (ENSG00000188822), score: 0.65 COL10A1collagen, type X, alpha 1 (ENSG00000123500), score: 0.51 CPSF6cleavage and polyadenylation specific factor 6, 68kDa (ENSG00000111605), score: -0.55 CTNNBIP1catenin, beta interacting protein 1 (ENSG00000178585), score: -0.57 CYB5R4cytochrome b5 reductase 4 (ENSG00000065615), score: 0.65 CYBRD1cytochrome b reductase 1 (ENSG00000071967), score: -0.72 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.56 DARS2aspartyl-tRNA synthetase 2, mitochondrial (ENSG00000117593), score: 0.49 DCTDdCMP deaminase (ENSG00000129187), score: -0.53 DCUN1D5DCN1, defective in cullin neddylation 1, domain containing 5 (S. cerevisiae) (ENSG00000137692), score: 0.49 DIMT1LDIM1 dimethyladenosine transferase 1-like (S. cerevisiae) (ENSG00000086189), score: -0.68 DPP8dipeptidyl-peptidase 8 (ENSG00000074603), score: 0.5 DTNBP1dystrobrevin binding protein 1 (ENSG00000047579), score: 0.49 ENGASEendo-beta-N-acetylglucosaminidase (ENSG00000167280), score: -0.54 ENTPD4ectonucleoside triphosphate diphosphohydrolase 4 (ENSG00000197217), score: 0.71 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: -0.77 FAM160B1family with sequence similarity 160, member B1 (ENSG00000151553), score: 0.54 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: 0.56 FAM199Xfamily with sequence similarity 199, X-linked (ENSG00000123575), score: -0.69 FAM36Afamily with sequence similarity 36, member A (ENSG00000203667), score: -0.56 FAM76Afamily with sequence similarity 76, member A (ENSG00000009780), score: 0.53 FBXL15F-box and leucine-rich repeat protein 15 (ENSG00000107872), score: -0.57 FBXL7F-box and leucine-rich repeat protein 7 (ENSG00000183580), score: -0.55 FBXO3F-box protein 3 (ENSG00000110429), score: 0.49 FBXW4F-box and WD repeat domain containing 4 (ENSG00000107829), score: -0.55 FBXW8F-box and WD repeat domain containing 8 (ENSG00000174989), score: 0.58 FKTNfukutin (ENSG00000106692), score: 0.48 FRS2fibroblast growth factor receptor substrate 2 (ENSG00000166225), score: 0.52 GABARAPL2GABA(A) receptor-associated protein-like 2 (ENSG00000034713), score: -0.63 GADD45Bgrowth arrest and DNA-damage-inducible, beta (ENSG00000099860), score: -0.56 GAPVD1GTPase activating protein and VPS9 domains 1 (ENSG00000165219), score: 0.48 GDE1glycerophosphodiester phosphodiesterase 1 (ENSG00000006007), score: 0.49 GGPS1geranylgeranyl diphosphate synthase 1 (ENSG00000152904), score: -0.59 GNA13guanine nucleotide binding protein (G protein), alpha 13 (ENSG00000120063), score: 0.49 GOLGA4golgin A4 (ENSG00000144674), score: 0.49 GPR180G protein-coupled receptor 180 (ENSG00000152749), score: 0.77 GXYLT1glucoside xylosyltransferase 1 (ENSG00000151233), score: 0.63 H1FXH1 histone family, member X (ENSG00000184897), score: -0.86 HDAC8histone deacetylase 8 (ENSG00000147099), score: -0.56 HDDC2HD domain containing 2 (ENSG00000111906), score: -0.71 HINFPhistone H4 transcription factor (ENSG00000172273), score: -0.71 HIPK3homeodomain interacting protein kinase 3 (ENSG00000110422), score: 0.6 HMBOX1homeobox containing 1 (ENSG00000147421), score: 0.51 HNRNPRheterogeneous nuclear ribonucleoprotein R (ENSG00000125944), score: -0.67 IGHMBP2immunoglobulin mu binding protein 2 (ENSG00000132740), score: -0.56 JAK2Janus kinase 2 (ENSG00000096968), score: 0.48 KCNRGpotassium channel regulator (ENSG00000198553), score: -0.53 KIAA0100KIAA0100 (ENSG00000007202), score: 0.5 KIAA2018KIAA2018 (ENSG00000176542), score: 0.53 LCLAT1lysocardiolipin acyltransferase 1 (ENSG00000172954), score: 0.59 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: 0.64 LOC100294349similar to Parkinson disease 7 domain containing 1 (ENSG00000177225), score: -0.53 LPGAT1lysophosphatidylglycerol acyltransferase 1 (ENSG00000123684), score: 0.59 LRSAM1leucine rich repeat and sterile alpha motif containing 1 (ENSG00000148356), score: -0.56 LRTM1leucine-rich repeats and transmembrane domains 1 (ENSG00000144771), score: 0.51 MAP1Dmethionine aminopeptidase 1D (ENSG00000172878), score: 0.65 MAP3K14mitogen-activated protein kinase kinase kinase 14 (ENSG00000006062), score: -0.53 MARCH5membrane-associated ring finger (C3HC4) 5 (ENSG00000198060), score: 0.66 MARCH6membrane-associated ring finger (C3HC4) 6 (ENSG00000145495), score: 0.48 MCM3APminichromosome maintenance complex component 3 associated protein (ENSG00000160294), score: -0.7 MDM4Mdm4 p53 binding protein homolog (mouse) (ENSG00000198625), score: 0.51 MEAF6MYST/Esa1-associated factor 6 (ENSG00000163875), score: -0.6 MED28mediator complex subunit 28 (ENSG00000118579), score: 0.8 METT10Dmethyltransferase 10 domain containing (ENSG00000127804), score: -0.59 MFN1mitofusin 1 (ENSG00000171109), score: 0.6 MIA3melanoma inhibitory activity family, member 3 (ENSG00000154305), score: 0.59 MICAL1microtubule associated monoxygenase, calponin and LIM domain containing 1 (ENSG00000135596), score: -0.55 MINPP1multiple inositol-polyphosphate phosphatase 1 (ENSG00000107789), score: 0.51 MIPOL1mirror-image polydactyly 1 (ENSG00000151338), score: 0.63 MPP6membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) (ENSG00000105926), score: 0.52 MRPL21mitochondrial ribosomal protein L21 (ENSG00000197345), score: -0.59 MRPS11mitochondrial ribosomal protein S11 (ENSG00000181991), score: -0.61 MTMR6myotubularin related protein 6 (ENSG00000139505), score: 0.6 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: -0.55 NAA35N(alpha)-acetyltransferase 35, NatC auxiliary subunit (ENSG00000135040), score: 0.63 NCBP1nuclear cap binding protein subunit 1, 80kDa (ENSG00000136937), score: 0.58 ND3NADH dehydrogenase, subunit 3 (complex I) (ENSG00000198840), score: -0.67 NEDD4neural precursor cell expressed, developmentally down-regulated 4 (ENSG00000069869), score: 0.88 NEIL1nei endonuclease VIII-like 1 (E. coli) (ENSG00000140398), score: -0.56 NSL1NSL1, MIND kinetochore complex component, homolog (S. cerevisiae) (ENSG00000117697), score: -0.86 NSUN2NOP2/Sun domain family, member 2 (ENSG00000037474), score: 0.49 NTHL1nth endonuclease III-like 1 (E. coli) (ENSG00000065057), score: -0.64 NUDCD1NudC domain containing 1 (ENSG00000120526), score: 0.49 NUDT19nudix (nucleoside diphosphate linked moiety X)-type motif 19 (ENSG00000213965), score: 0.54 NUDT4nudix (nucleoside diphosphate linked moiety X)-type motif 4 (ENSG00000173598), score: 0.52 OSGIN2oxidative stress induced growth inhibitor family member 2 (ENSG00000164823), score: -0.55 PAK1IP1PAK1 interacting protein 1 (ENSG00000111845), score: 0.63 PALB2partner and localizer of BRCA2 (ENSG00000083093), score: -0.63 PAQR9progestin and adipoQ receptor family member IX (ENSG00000188582), score: 0.49 PEX10peroxisomal biogenesis factor 10 (ENSG00000157911), score: -0.73 PGAP3post-GPI attachment to proteins 3 (ENSG00000161395), score: -0.64 PKD2L2polycystic kidney disease 2-like 2 (ENSG00000078795), score: 0.55 PLEKHN1pleckstrin homology domain containing, family N member 1 (ENSG00000187583), score: 0.78 POLR3Gpolymerase (RNA) III (DNA directed) polypeptide G (32kD) (ENSG00000113356), score: 0.5 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: -0.57 PPM1Bprotein phosphatase, Mg2+/Mn2+ dependent, 1B (ENSG00000138032), score: 0.59 PPP1R7protein phosphatase 1, regulatory (inhibitor) subunit 7 (ENSG00000115685), score: -0.62 PQLC1PQ loop repeat containing 1 (ENSG00000122490), score: -0.56 PQLC3PQ loop repeat containing 3 (ENSG00000162976), score: -0.55 PROX2prospero homeobox 2 (ENSG00000119608), score: 0.57 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: -0.59 PRSSL1protease, serine-like 1 (ENSG00000185198), score: 0.52 PSMD12proteasome (prosome, macropain) 26S subunit, non-ATPase, 12 (ENSG00000197170), score: 0.63 PSMG3proteasome (prosome, macropain) assembly chaperone 3 (ENSG00000157778), score: -0.65 PSTPIP2proline-serine-threonine phosphatase interacting protein 2 (ENSG00000152229), score: 0.56 PTPLAD2protein tyrosine phosphatase-like A domain containing 2 (ENSG00000188921), score: 0.48 PUS10pseudouridylate synthase 10 (ENSG00000162927), score: 0.54 RAB22ARAB22A, member RAS oncogene family (ENSG00000124209), score: 0.53 RAD51RAD51 homolog (RecA homolog, E. coli) (S. cerevisiae) (ENSG00000051180), score: 0.49 RFESDRieske (Fe-S) domain containing (ENSG00000175449), score: 0.65 RFWD3ring finger and WD repeat domain 3 (ENSG00000168411), score: 0.56 RNF139ring finger protein 139 (ENSG00000170881), score: 0.76 RPS19BP1ribosomal protein S19 binding protein 1 (ENSG00000187051), score: -0.66 RPUSD2RNA pseudouridylate synthase domain containing 2 (ENSG00000166133), score: -0.62 RRAGCRas-related GTP binding C (ENSG00000116954), score: 0.52 SAP30BPSAP30 binding protein (ENSG00000161526), score: -0.65 SEC23IPSEC23 interacting protein (ENSG00000107651), score: 0.6 SEC31BSEC31 homolog B (S. cerevisiae) (ENSG00000075826), score: -0.67 SERINC3serine incorporator 3 (ENSG00000132824), score: 0.52 SGSM3small G protein signaling modulator 3 (ENSG00000100359), score: -0.57 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: -0.68 SLC25A29solute carrier family 25, member 29 (ENSG00000197119), score: -0.55 SLC6A6solute carrier family 6 (neurotransmitter transporter, taurine), member 6 (ENSG00000131389), score: 0.49 SMARCA1SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 1 (ENSG00000102038), score: -0.73 SMC5structural maintenance of chromosomes 5 (ENSG00000198887), score: 0.48 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: 0.51 SNX11sorting nexin 11 (ENSG00000002919), score: -0.57 SNX17sorting nexin 17 (ENSG00000115234), score: -0.59 SSBP1single-stranded DNA binding protein 1 (ENSG00000106028), score: -0.63 SSPNsarcospan (Kras oncogene-associated gene) (ENSG00000123096), score: 0.81 STK17Bserine/threonine kinase 17b (ENSG00000081320), score: 0.49 TAF1ATATA box binding protein (TBP)-associated factor, RNA polymerase I, A, 48kDa (ENSG00000143498), score: 0.61 TBCELtubulin folding cofactor E-like (ENSG00000154114), score: 0.59 TCP11L1t-complex 11 (mouse)-like 1 (ENSG00000176148), score: -0.66 TELO2TEL2, telomere maintenance 2, homolog (S. cerevisiae) (ENSG00000100726), score: -0.62 TMBIM4transmembrane BAX inhibitor motif containing 4 (ENSG00000155957), score: 0.5 TMEM106Btransmembrane protein 106B (ENSG00000106460), score: 0.54 TMEM14Atransmembrane protein 14A (ENSG00000096092), score: -0.68 TMEM64transmembrane protein 64 (ENSG00000180694), score: 0.73 TMF1TATA element modulatory factor 1 (ENSG00000144747), score: 0.48 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: 0.56 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: 0.54 TOPORStopoisomerase I binding, arginine/serine-rich (ENSG00000197579), score: 0.48 TXNDC16thioredoxin domain containing 16 (ENSG00000087301), score: 0.55 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: 0.73 TYW3tRNA-yW synthesizing protein 3 homolog (S. cerevisiae) (ENSG00000162623), score: -0.56 UBE2Wubiquitin-conjugating enzyme E2W (putative) (ENSG00000104343), score: 0.62 UBLCP1ubiquitin-like domain containing CTD phosphatase 1 (ENSG00000164332), score: 0.55 UBR2ubiquitin protein ligase E3 component n-recognin 2 (ENSG00000024048), score: 0.61 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.48 USP47ubiquitin specific peptidase 47 (ENSG00000170242), score: 0.58 USP8ubiquitin specific peptidase 8 (ENSG00000138592), score: 0.49 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (ENSG00000197969), score: 0.53 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: 0.63 VPS45vacuolar protein sorting 45 homolog (S. cerevisiae) (ENSG00000136631), score: -0.54 WBP4WW domain binding protein 4 (formin binding protein 21) (ENSG00000120688), score: 0.54 WDR36WD repeat domain 36 (ENSG00000134987), score: 0.65 WDR89WD repeat domain 89 (ENSG00000140006), score: 0.89 XRCC4X-ray repair complementing defective repair in Chinese hamster cells 4 (ENSG00000152422), score: 0.51 YTHDF2YTH domain family, member 2 (ENSG00000198492), score: -0.54 ZBTB41zinc finger and BTB domain containing 41 (ENSG00000177888), score: 0.57 ZC3H7Azinc finger CCCH-type containing 7A (ENSG00000122299), score: 0.91 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: 0.54 ZNF750zinc finger protein 750 (ENSG00000141579), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_kd_m1_ca1 | hsa | kd | m | 1 |
| ptr_br_m1_ca1 | ptr | br | m | 1 |
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| ptr_br_m2_ca1 | ptr | br | m | 2 |
| ppa_br_f1_ca1 | ppa | br | f | 1 |
| ggo_br_m_ca1 | ggo | br | m | _ |
| mdo_ts_m2_ca1 | mdo | ts | m | 2 |
| mdo_ts_m1_ca1 | mdo | ts | m | 1 |
| mmu_cb_m1_ca1 | mmu | cb | m | 1 |
| mmu_br_m1_ca1 | mmu | br | m | 1 |
| mmu_br_m2_ca1 | mmu | br | m | 2 |
| mmu_br_f_ca1 | mmu | br | f | _ |
| mmu_cb_f_ca1 | mmu | cb | f | _ |
| mmu_cb_m2_ca1 | mmu | cb | m | 2 |
| mmu_ht_f_ca1 | mmu | ht | f | _ |
| mmu_ht_m1_ca1 | mmu | ht | m | 1 |
| mmu_kd_m1_ca1 | mmu | kd | m | 1 |
| mmu_lv_m1_ca1 | mmu | lv | m | 1 |
| mmu_kd_f_ca1 | mmu | kd | f | _ |
| mmu_ht_m2_ca1 | mmu | ht | m | 2 |
| mmu_kd_m2_ca1 | mmu | kd | m | 2 |
| mmu_ts_m1_ca1 | mmu | ts | m | 1 |
| mmu_lv_f_ca1 | mmu | lv | f | _ |
| mmu_lv_m2_ca1 | mmu | lv | m | 2 |
| mmu_ts_m2_ca1 | mmu | ts | m | 2 |