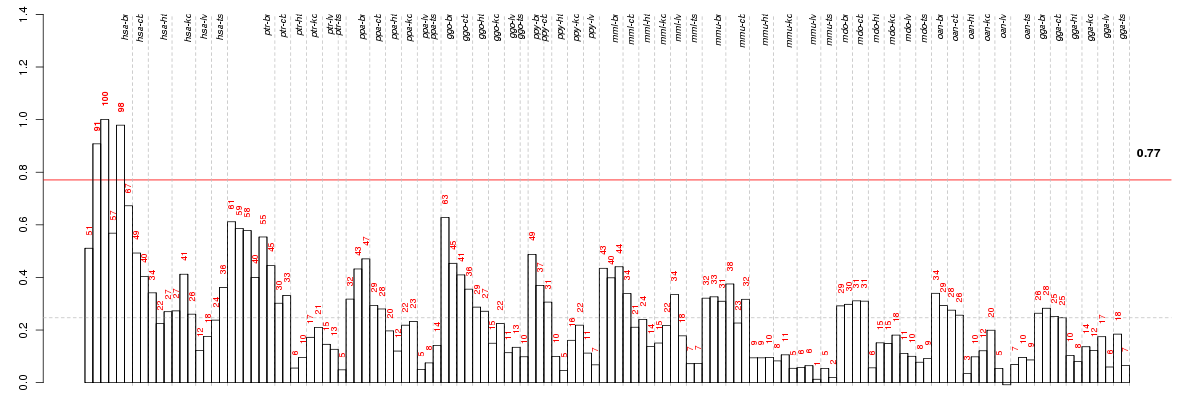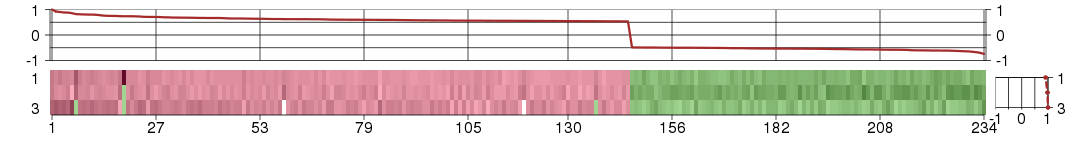

Under-expression is coded with green,
over-expression with red color.



molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
nucleic acid binding
Interacting selectively and non-covalently with any nucleic acid.
translation initiation factor activity
Functions in the initiation of ribosome-mediated translation of mRNA into a polypeptide.
binding
The selective, non-covalent, often stoichiometric, interaction of a molecule with one or more specific sites on another molecule.
translation factor activity, nucleic acid binding
Functions during translation by binding nucleic acids during polypeptide synthesis at the ribosome.
all
NA
ABCE1ATP-binding cassette, sub-family E (OABP), member 1 (ENSG00000164163), score: -0.6 ABL1c-abl oncogene 1, non-receptor tyrosine kinase (ENSG00000097007), score: 0.56 ADAMTS14ADAM metallopeptidase with thrombospondin type 1 motif, 14 (ENSG00000138316), score: 0.8 ADARB2adenosine deaminase, RNA-specific, B2 (ENSG00000185736), score: 0.65 AKAP9A kinase (PRKA) anchor protein (yotiao) 9 (ENSG00000127914), score: -0.58 ALKBH8alkB, alkylation repair homolog 8 (E. coli) (ENSG00000137760), score: -0.54 ANO4anoctamin 4 (ENSG00000151572), score: 0.54 AP3M1adaptor-related protein complex 3, mu 1 subunit (ENSG00000185009), score: -0.49 ARFIP1ADP-ribosylation factor interacting protein 1 (ENSG00000164144), score: -0.5 ARHGAP1Rho GTPase activating protein 1 (ENSG00000175220), score: 0.6 ARHGAP22Rho GTPase activating protein 22 (ENSG00000128805), score: 0.6 ARRDC2arrestin domain containing 2 (ENSG00000105643), score: 0.57 ATL2atlastin GTPase 2 (ENSG00000119787), score: -0.74 ATP11AATPase, class VI, type 11A (ENSG00000068650), score: 0.54 BACE1beta-site APP-cleaving enzyme 1 (ENSG00000186318), score: 0.55 BCAS3breast carcinoma amplified sequence 3 (ENSG00000141376), score: 0.64 C12orf45chromosome 12 open reading frame 45 (ENSG00000151131), score: -0.51 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: 0.56 C16orf70chromosome 16 open reading frame 70 (ENSG00000125149), score: 0.6 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: 0.73 C5orf41chromosome 5 open reading frame 41 (ENSG00000164463), score: -0.51 CALHM1calcium homeostasis modulator 1 (ENSG00000185933), score: 0.82 CAPN9calpain 9 (ENSG00000135773), score: 0.88 CCDC117coiled-coil domain containing 117 (ENSG00000159873), score: -0.53 CDC42BPBCDC42 binding protein kinase beta (DMPK-like) (ENSG00000198752), score: 0.6 CDH4cadherin 4, type 1, R-cadherin (retinal) (ENSG00000179242), score: 0.56 CEBPZCCAAT/enhancer binding protein (C/EBP), zeta (ENSG00000115816), score: -0.5 CERCAMcerebral endothelial cell adhesion molecule (ENSG00000167123), score: 1 CETN3centrin, EF-hand protein, 3 (ENSG00000153140), score: -0.53 CLCN7chloride channel 7 (ENSG00000103249), score: 0.6 CLDND1claudin domain containing 1 (ENSG00000080822), score: 0.9 CNTN2contactin 2 (axonal) (ENSG00000184144), score: 0.62 COG1component of oligomeric golgi complex 1 (ENSG00000166685), score: 0.55 COMMD10COMM domain containing 10 (ENSG00000145781), score: -0.53 CPNE2copine II (ENSG00000140848), score: 0.67 CSRP1cysteine and glycine-rich protein 1 (ENSG00000159176), score: 0.58 CTNNAL1catenin (cadherin-associated protein), alpha-like 1 (ENSG00000119326), score: -0.5 CTRLchymotrypsin-like (ENSG00000141086), score: 0.62 CUL5cullin 5 (ENSG00000166266), score: -0.5 DAAM2dishevelled associated activator of morphogenesis 2 (ENSG00000146122), score: 0.71 DDX46DEAD (Asp-Glu-Ala-Asp) box polypeptide 46 (ENSG00000145833), score: -0.5 DDX52DEAD (Asp-Glu-Ala-Asp) box polypeptide 52 (ENSG00000141141), score: -0.57 DFNB31deafness, autosomal recessive 31 (ENSG00000095397), score: 0.55 DGCR2DiGeorge syndrome critical region gene 2 (ENSG00000070413), score: 0.55 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.51 DNMBPdynamin binding protein (ENSG00000107554), score: -0.61 DOHHdeoxyhypusine hydroxylase/monooxygenase (ENSG00000129932), score: 0.66 DSCAML1Down syndrome cell adhesion molecule like 1 (ENSG00000177103), score: 0.6 DSELdermatan sulfate epimerase-like (ENSG00000171451), score: 0.56 DYNC1I2dynein, cytoplasmic 1, intermediate chain 2 (ENSG00000077380), score: 0.68 EIF2S1eukaryotic translation initiation factor 2, subunit 1 alpha, 35kDa (ENSG00000134001), score: -0.51 EIF3Eeukaryotic translation initiation factor 3, subunit E (ENSG00000104408), score: -0.5 EIF3Meukaryotic translation initiation factor 3, subunit M (ENSG00000149100), score: -0.53 EIF4Heukaryotic translation initiation factor 4H (ENSG00000106682), score: 0.59 EIF5Beukaryotic translation initiation factor 5B (ENSG00000158417), score: -0.54 ELOVL1elongation of very long chain fatty acids (FEN1/Elo2, SUR4/Elo3, yeast)-like 1 (ENSG00000066322), score: 0.74 ENPP2ectonucleotide pyrophosphatase/phosphodiesterase 2 (ENSG00000136960), score: 0.57 EPB41erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) (ENSG00000159023), score: -0.54 EPN2epsin 2 (ENSG00000072134), score: 0.55 ERMNermin, ERM-like protein (ENSG00000136541), score: 0.54 EVI2Aecotropic viral integration site 2A (ENSG00000126860), score: 0.67 EXOC7exocyst complex component 7 (ENSG00000182473), score: 0.61 FA2Hfatty acid 2-hydroxylase (ENSG00000103089), score: 0.65 FAM102Afamily with sequence similarity 102, member A (ENSG00000167106), score: 0.78 FAM124Afamily with sequence similarity 124A (ENSG00000150510), score: 0.81 FAM171A1family with sequence similarity 171, member A1 (ENSG00000148468), score: 0.54 FAM175Bfamily with sequence similarity 175, member B (ENSG00000165660), score: -0.64 FAM69Cfamily with sequence similarity 69, member C (ENSG00000187773), score: 0.64 FASTKD2FAST kinase domains 2 (ENSG00000118246), score: -0.53 FGF22fibroblast growth factor 22 (ENSG00000070388), score: 0.62 FKBP15FK506 binding protein 15, 133kDa (ENSG00000119321), score: 0.59 FNBP1Lformin binding protein 1-like (ENSG00000137942), score: -0.61 FZD6frizzled homolog 6 (Drosophila) (ENSG00000164930), score: -0.54 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.62 GLDNgliomedin (ENSG00000186417), score: 0.79 GNL2guanine nucleotide binding protein-like 2 (nucleolar) (ENSG00000134697), score: -0.58 GNPTGN-acetylglucosamine-1-phosphate transferase, gamma subunit (ENSG00000090581), score: 0.54 GOLGA4golgin A4 (ENSG00000144674), score: -0.62 GORABgolgin, RAB6-interacting (ENSG00000120370), score: -0.57 GPR123G protein-coupled receptor 123 (ENSG00000197177), score: 0.71 GRID1glutamate receptor, ionotropic, delta 1 (ENSG00000182771), score: 0.59 GRM3glutamate receptor, metabotropic 3 (ENSG00000198822), score: 0.55 GTF2A2general transcription factor IIA, 2, 12kDa (ENSG00000140307), score: -0.5 GTF2E2general transcription factor IIE, polypeptide 2, beta 34kDa (ENSG00000197265), score: -0.57 HDAC11histone deacetylase 11 (ENSG00000163517), score: 0.57 HGShepatocyte growth factor-regulated tyrosine kinase substrate (ENSG00000185359), score: 0.62 HHIPhedgehog interacting protein (ENSG00000164161), score: 0.56 HHIPL1HHIP-like 1 (ENSG00000182218), score: 0.6 HOOK1hook homolog 1 (Drosophila) (ENSG00000134709), score: -0.52 ICOSLGinducible T-cell co-stimulator ligand (ENSG00000160223), score: 0.68 IL1RAPL1interleukin 1 receptor accessory protein-like 1 (ENSG00000169306), score: 0.62 ING3inhibitor of growth family, member 3 (ENSG00000071243), score: -0.52 INTS8integrator complex subunit 8 (ENSG00000164941), score: -0.54 ITPK1inositol 1,3,4-triphosphate 5/6 kinase (ENSG00000100605), score: 0.63 JAM3junctional adhesion molecule 3 (ENSG00000166086), score: 0.68 KCNF1potassium voltage-gated channel, subfamily F, member 1 (ENSG00000162975), score: 0.56 KCNH8potassium voltage-gated channel, subfamily H (eag-related), member 8 (ENSG00000183960), score: 0.92 KCNMB4potassium large conductance calcium-activated channel, subfamily M, beta member 4 (ENSG00000135643), score: 0.56 KIAA1429KIAA1429 (ENSG00000164944), score: -0.55 KIF13Bkinesin family member 13B (ENSG00000197892), score: 0.55 KLHL26kelch-like 26 (Drosophila) (ENSG00000167487), score: 0.56 L3MBTL2l(3)mbt-like 2 (Drosophila) (ENSG00000100395), score: 0.54 LAPTM4Alysosomal protein transmembrane 4 alpha (ENSG00000068697), score: -0.56 LGR5leucine-rich repeat-containing G protein-coupled receptor 5 (ENSG00000139292), score: 0.6 LIN7Clin-7 homolog C (C. elegans) (ENSG00000148943), score: -0.53 LIX1Lix1 homolog (chicken) (ENSG00000145721), score: 0.69 LOC100131693eukaryotic translation initiation factor 4E pseudogene (ENSG00000151247), score: -0.58 LOC100294412similar to KIAA0655 protein (ENSG00000130787), score: 0.67 LOC652522similar to Werner syndrome protein (ENSG00000165392), score: -0.51 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: 0.56 LTV1LTV1 homolog (S. cerevisiae) (ENSG00000135521), score: -0.62 LZTS2leucine zipper, putative tumor suppressor 2 (ENSG00000107816), score: 0.59 MALmal, T-cell differentiation protein (ENSG00000172005), score: 0.54 MAP3K1mitogen-activated protein kinase kinase kinase 1 (ENSG00000095015), score: -0.52 MAP4K3mitogen-activated protein kinase kinase kinase kinase 3 (ENSG00000011566), score: -0.58 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: -0.51 MATN1matrilin 1, cartilage matrix protein (ENSG00000162510), score: 0.67 METTL14methyltransferase like 14 (ENSG00000145388), score: -0.49 MGRN1mahogunin, ring finger 1 (ENSG00000102858), score: 0.55 MNAT1menage a trois homolog 1, cyclin H assembly factor (Xenopus laevis) (ENSG00000020426), score: -0.6 MOCS1molybdenum cofactor synthesis 1 (ENSG00000124615), score: 0.57 MPLmyeloproliferative leukemia virus oncogene (ENSG00000117400), score: 0.58 MRPL15mitochondrial ribosomal protein L15 (ENSG00000137547), score: -0.5 MRPL50mitochondrial ribosomal protein L50 (ENSG00000136897), score: -0.55 MSGN1mesogenin 1 (ENSG00000151379), score: 0.74 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: -0.59 MTMR10myotubularin related protein 10 (ENSG00000166912), score: 0.55 MTPAPmitochondrial poly(A) polymerase (ENSG00000107951), score: -0.52 MVDmevalonate (diphospho) decarboxylase (ENSG00000167508), score: 0.55 MYNNmyoneurin (ENSG00000085274), score: -0.57 MYO10myosin X (ENSG00000145555), score: 0.56 MYO9Bmyosin IXB (ENSG00000099331), score: 0.71 NDST1N-deacetylase/N-sulfotransferase (heparan glucosaminyl) 1 (ENSG00000070614), score: 0.55 NINJ2ninjurin 2 (ENSG00000171840), score: 0.56 NIPA1non imprinted in Prader-Willi/Angelman syndrome 1 (ENSG00000170113), score: 0.59 NIPAL4NIPA-like domain containing 4 (ENSG00000172548), score: 0.75 NOP58NOP58 ribonucleoprotein homolog (yeast) (ENSG00000055044), score: -0.52 NOTCH1notch 1 (ENSG00000148400), score: 0.55 NPC1Niemann-Pick disease, type C1 (ENSG00000141458), score: 0.56 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: -0.5 NUCB2nucleobindin 2 (ENSG00000070081), score: -0.54 OPN4opsin 4 (ENSG00000122375), score: 0.65 PACS2phosphofurin acidic cluster sorting protein 2 (ENSG00000179364), score: 0.62 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.69 PEX13peroxisomal biogenesis factor 13 (ENSG00000162928), score: -0.56 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: 0.6 PHLDB1pleckstrin homology-like domain, family B, member 1 (ENSG00000019144), score: 0.59 PHLPP1PH domain and leucine rich repeat protein phosphatase 1 (ENSG00000081913), score: 0.62 PIGBphosphatidylinositol glycan anchor biosynthesis, class B (ENSG00000069943), score: -0.55 PIP4K2Aphosphatidylinositol-5-phosphate 4-kinase, type II, alpha (ENSG00000150867), score: 0.75 PLXNB1plexin B1 (ENSG00000164050), score: 0.58 PMP2peripheral myelin protein 2 (ENSG00000147588), score: 0.56 POLD3polymerase (DNA-directed), delta 3, accessory subunit (ENSG00000077514), score: -0.5 PPIGpeptidylprolyl isomerase G (cyclophilin G) (ENSG00000138398), score: -0.53 PREX1phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 1 (ENSG00000124126), score: 0.8 PRR5Lproline rich 5 like (ENSG00000135362), score: 0.76 PRRG1proline rich Gla (G-carboxyglutamic acid) 1 (ENSG00000130962), score: 0.59 PTK2PTK2 protein tyrosine kinase 2 (ENSG00000169398), score: 0.75 RAB40CRAB40C, member RAS oncogene family (ENSG00000197562), score: 0.57 RANGAP1Ran GTPase activating protein 1 (ENSG00000100401), score: 0.62 RAPGEF5Rap guanine nucleotide exchange factor (GEF) 5 (ENSG00000136237), score: 0.65 RASAL1RAS protein activator like 1 (GAP1 like) (ENSG00000111344), score: 0.68 RBP3retinol binding protein 3, interstitial (ENSG00000107618), score: 0.86 RGS11regulator of G-protein signaling 11 (ENSG00000076344), score: 0.54 RHOrhodopsin (ENSG00000163914), score: 0.54 RHOUras homolog gene family, member U (ENSG00000116574), score: 0.54 RIOK2RIO kinase 2 (yeast) (ENSG00000058729), score: -0.67 ROBO3roundabout, axon guidance receptor, homolog 3 (Drosophila) (ENSG00000154134), score: 0.63 ROCK1Rho-associated, coiled-coil containing protein kinase 1 (ENSG00000067900), score: -0.55 RPF1ribosome production factor 1 homolog (S. cerevisiae) (ENSG00000117133), score: -0.7 RRP15ribosomal RNA processing 15 homolog (S. cerevisiae) (ENSG00000067533), score: -0.58 RS1retinoschisin 1 (ENSG00000102104), score: 0.74 RSPO4R-spondin family, member 4 (ENSG00000101282), score: 0.58 SECISBP2LSECIS binding protein 2-like (ENSG00000138593), score: 0.54 SEMA6Asema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A (ENSG00000092421), score: 0.71 SETD6SET domain containing 6 (ENSG00000103037), score: -0.53 SFRS12splicing factor, arginine/serine-rich 12 (ENSG00000153914), score: -0.6 SH3GLB2SH3-domain GRB2-like endophilin B2 (ENSG00000148341), score: 0.57 SH3PXD2BSH3 and PX domains 2B (ENSG00000174705), score: 0.54 SIK3SIK family kinase 3 (ENSG00000160584), score: 0.59 SLC31A2solute carrier family 31 (copper transporters), member 2 (ENSG00000136867), score: 0.54 SLC44A1solute carrier family 44, member 1 (ENSG00000070214), score: 0.71 SLC5A11solute carrier family 5 (sodium/glucose cotransporter), member 11 (ENSG00000158865), score: 0.67 SMEK1SMEK homolog 1, suppressor of mek1 (Dictyostelium) (ENSG00000100796), score: -0.57 SMNDC1survival motor neuron domain containing 1 (ENSG00000119953), score: -0.61 SNX13sorting nexin 13 (ENSG00000071189), score: -0.49 SOX8SRY (sex determining region Y)-box 8 (ENSG00000005513), score: 0.67 SREBF2sterol regulatory element binding transcription factor 2 (ENSG00000198911), score: 0.53 STARD3StAR-related lipid transfer (START) domain containing 3 (ENSG00000131748), score: 0.67 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: -0.49 SUZ12suppressor of zeste 12 homolog (Drosophila) (ENSG00000178691), score: -0.66 SYNJ2synaptojanin 2 (ENSG00000078269), score: 0.63 TAF12TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa (ENSG00000120656), score: -0.54 TBC1D2TBC1 domain family, member 2 (ENSG00000095383), score: 0.68 TBC1D23TBC1 domain family, member 23 (ENSG00000036054), score: -0.57 TBC1D5TBC1 domain family, member 5 (ENSG00000131374), score: 0.53 TBCEtubulin folding cofactor E (ENSG00000116957), score: -0.49 TDRD3tudor domain containing 3 (ENSG00000083544), score: -0.61 THOC2THO complex 2 (ENSG00000125676), score: -0.61 TJAP1tight junction associated protein 1 (peripheral) (ENSG00000137221), score: 0.73 TMC6transmembrane channel-like 6 (ENSG00000141524), score: 0.88 TMCC3transmembrane and coiled-coil domain family 3 (ENSG00000057704), score: 0.57 TMEM144transmembrane protein 144 (ENSG00000164124), score: 0.81 TMEM175transmembrane protein 175 (ENSG00000127419), score: 0.74 TMEM179transmembrane protein 179 (ENSG00000189203), score: 0.56 TMEM184Btransmembrane protein 184B (ENSG00000198792), score: 0.6 TMTC2transmembrane and tetratricopeptide repeat containing 2 (ENSG00000179104), score: 0.63 TOMM70Atranslocase of outer mitochondrial membrane 70 homolog A (S. cerevisiae) (ENSG00000154174), score: -0.6 TOP1topoisomerase (DNA) I (ENSG00000198900), score: -0.53 TPPPtubulin polymerization promoting protein (ENSG00000171368), score: 0.55 TRMT61AtRNA methyltransferase 61 homolog A (S. cerevisiae) (ENSG00000166166), score: 0.63 TRPM7transient receptor potential cation channel, subfamily M, member 7 (ENSG00000092439), score: -0.56 TSPAN15tetraspanin 15 (ENSG00000099282), score: 0.65 TTC15tetratricopeptide repeat domain 15 (ENSG00000171853), score: 0.54 TXNDC9thioredoxin domain containing 9 (ENSG00000115514), score: -0.53 UNC5Bunc-5 homolog B (C. elegans) (ENSG00000107731), score: 0.58 USO1USO1 vesicle docking protein homolog (yeast) (ENSG00000138768), score: -0.49 USP54ubiquitin specific peptidase 54 (ENSG00000166348), score: 0.62 UTP11LUTP11-like, U3 small nucleolar ribonucleoprotein, (yeast) (ENSG00000183520), score: -0.63 VPS13Avacuolar protein sorting 13 homolog A (S. cerevisiae) (ENSG00000197969), score: -0.49 VPS26Avacuolar protein sorting 26 homolog A (S. pombe) (ENSG00000122958), score: -0.5 WBP1WW domain binding protein 1 (ENSG00000115274), score: 0.57 WDR36WD repeat domain 36 (ENSG00000134987), score: -0.6 WDR43WD repeat domain 43 (ENSG00000163811), score: -0.55 WEE1WEE1 homolog (S. pombe) (ENSG00000166483), score: -0.53 WIPF1WAS/WASL interacting protein family, member 1 (ENSG00000115935), score: 0.56 XRN15'-3' exoribonuclease 1 (ENSG00000114127), score: -0.58 YME1L1YME1-like 1 (S. cerevisiae) (ENSG00000136758), score: -0.64 ZBTB17zinc finger and BTB domain containing 17 (ENSG00000116809), score: 0.68 ZCCHC8zinc finger, CCHC domain containing 8 (ENSG00000033030), score: -0.5 ZEB2zinc finger E-box binding homeobox 2 (ENSG00000169554), score: 0.64 ZFAND6zinc finger, AN1-type domain 6 (ENSG00000086666), score: -0.55 ZNF276zinc finger protein 276 (ENSG00000158805), score: 0.61 ZNF536zinc finger protein 536 (ENSG00000198597), score: 0.64
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| hsa_br_m3_ca1 | hsa | br | m | 3 |
| hsa_br_m7_ca1 | hsa | br | m | 7 |
| hsa_br_m6_ca1 | hsa | br | m | 6 |