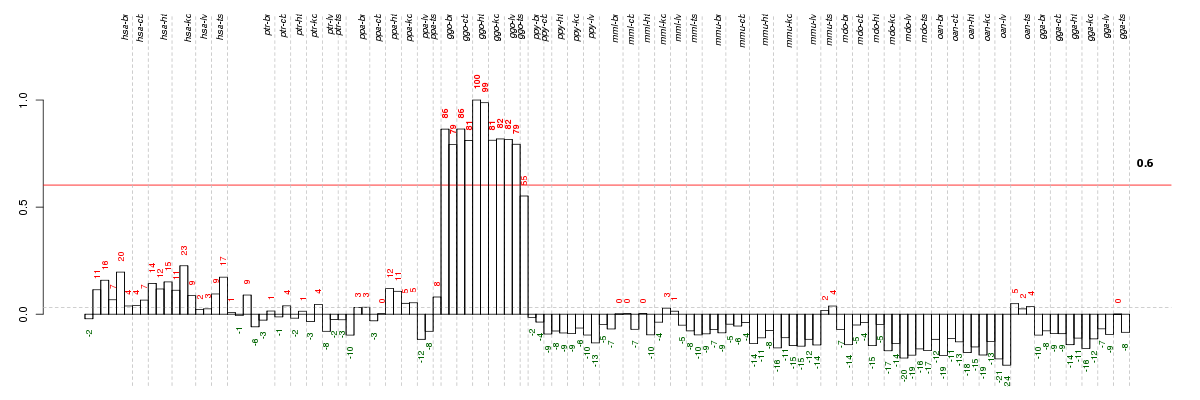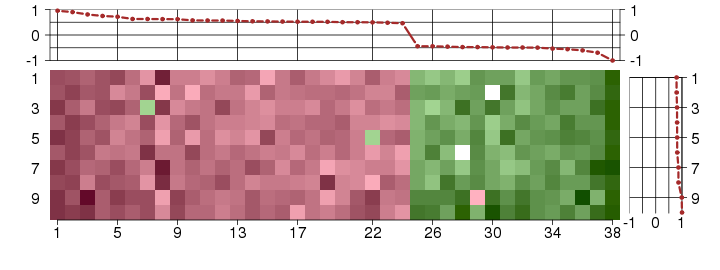

Under-expression is coded with green,
over-expression with red color.



AAGABalpha- and gamma-adaptin binding protein (ENSG00000103591), score: -0.47 AHCTF1AT hook containing transcription factor 1 (ENSG00000153207), score: -0.6 ANKRD54ankyrin repeat domain 54 (ENSG00000100124), score: -0.56 ARCN1archain 1 (ENSG00000095139), score: -0.46 BEST1bestrophin 1 (ENSG00000167995), score: 0.52 C8orf45chromosome 8 open reading frame 45 (ENSG00000178460), score: 0.95 CACNA2D4calcium channel, voltage-dependent, alpha 2/delta subunit 4 (ENSG00000151062), score: 0.9 CCDC111coiled-coil domain containing 111 (ENSG00000164306), score: 0.46 CCDC51coiled-coil domain containing 51 (ENSG00000164051), score: 0.53 COL8A1collagen, type VIII, alpha 1 (ENSG00000144810), score: 0.5 CPSF4Lcleavage and polyadenylation specific factor 4-like (ENSG00000187959), score: 0.63 CUL1cullin 1 (ENSG00000055130), score: -1 DCTdopachrome tautomerase (dopachrome delta-isomerase, tyrosine-related protein 2) (ENSG00000080166), score: 0.52 DDX55DEAD (Asp-Glu-Ala-Asp) box polypeptide 55 (ENSG00000111364), score: 0.52 DNAJC24DnaJ (Hsp40) homolog, subfamily C, member 24 (ENSG00000170946), score: -0.44 DVL3dishevelled, dsh homolog 3 (Drosophila) (ENSG00000161202), score: 0.54 EMILIN3elastin microfibril interfacer 3 (ENSG00000183798), score: 0.63 EPT1ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) (ENSG00000138018), score: -0.49 FAM59Bfamily with sequence similarity 59, member B (ENSG00000157833), score: 0.62 HKDC1hexokinase domain containing 1 (ENSG00000156510), score: 0.72 IP6K3inositol hexakisphosphate kinase 3 (ENSG00000161896), score: 0.5 ITGA11integrin, alpha 11 (ENSG00000137809), score: 0.75 KIF15kinesin family member 15 (ENSG00000163808), score: 0.55 LOC100289938similar to cytoplasmic tRNA 2-thiolation protein 2 (ENSG00000174177), score: 0.53 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: 0.57 MFSD2Bmajor facilitator superfamily domain containing 2B (ENSG00000205639), score: 0.62 NDUFB2NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 2, 8kDa (ENSG00000090266), score: -0.47 NME7non-metastatic cells 7, protein expressed in (nucleoside-diphosphate kinase) (ENSG00000143156), score: -0.53 PCID2PCI domain containing 2 (ENSG00000126226), score: 0.48 PDAP1PDGFA associated protein 1 (ENSG00000106244), score: 0.5 PDE6Bphosphodiesterase 6B, cGMP-specific, rod, beta (ENSG00000133256), score: 0.57 PPIL3peptidylprolyl isomerase (cyclophilin)-like 3 (ENSG00000240344), score: -0.48 PTCD1pentatricopeptide repeat domain 1 (ENSG00000106246), score: 0.81 RALAv-ral simian leukemia viral oncogene homolog A (ras related) (ENSG00000006451), score: -0.69 RECQL4RecQ protein-like 4 (ENSG00000160957), score: 0.57 TTC33tetratricopeptide repeat domain 33 (ENSG00000113638), score: -0.44 UBR7ubiquitin protein ligase E3 component n-recognin 7 (putative) (ENSG00000012963), score: -0.49 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: -0.5
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| ggo_br_f_ca1 | ggo | br | f | _ |
| ggo_lv_f_ca1 | ggo | lv | f | _ |
| ggo_cb_f_ca1 | ggo | cb | f | _ |
| ggo_kd_m_ca1 | ggo | kd | m | _ |
| ggo_lv_m_ca1 | ggo | lv | m | _ |
| ggo_kd_f_ca1 | ggo | kd | f | _ |
| ggo_br_m_ca1 | ggo | br | m | _ |
| ggo_cb_m_ca1 | ggo | cb | m | _ |
| ggo_ht_f_ca1 | ggo | ht | f | _ |
| ggo_ht_m_ca1 | ggo | ht | m | _ |