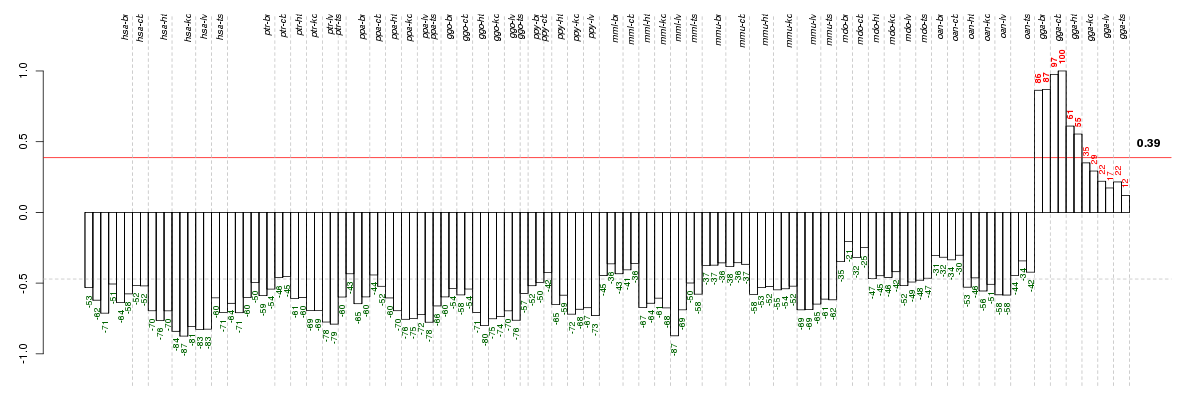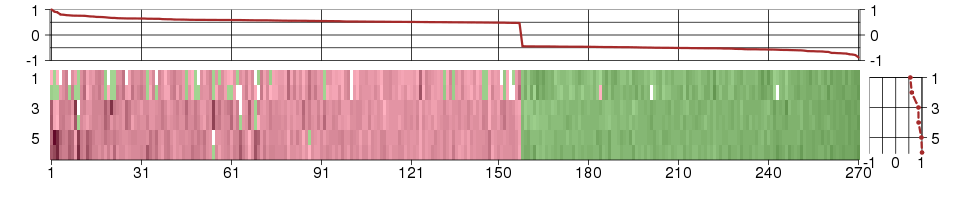

Under-expression is coded with green,
over-expression with red color.



AAK1AP2 associated kinase 1 (ENSG00000115977), score: 0.49 ADAMTS3ADAM metallopeptidase with thrombospondin type 1 motif, 3 (ENSG00000156140), score: 0.48 ADAT1adenosine deaminase, tRNA-specific 1 (ENSG00000065457), score: 0.65 ADAT2adenosine deaminase, tRNA-specific 2, TAD2 homolog (S. cerevisiae) (ENSG00000189007), score: 0.56 ADNPactivity-dependent neuroprotector homeobox (ENSG00000101126), score: 0.53 ALG5asparagine-linked glycosylation 5, dolichyl-phosphate beta-glucosyltransferase homolog (S. cerevisiae) (ENSG00000120697), score: -0.48 ALX4ALX homeobox 4 (ENSG00000052850), score: 0.69 AMHanti-Mullerian hormone (ENSG00000104899), score: 0.55 AMOTangiomotin (ENSG00000126016), score: 0.57 ANGPT2angiopoietin 2 (ENSG00000091879), score: 0.8 ANKRA2ankyrin repeat, family A (RFXANK-like), 2 (ENSG00000164331), score: 0.54 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.49 ANKRD44ankyrin repeat domain 44 (ENSG00000065413), score: 0.65 ANO5anoctamin 5 (ENSG00000171714), score: 0.53 ARFGAP2ADP-ribosylation factor GTPase activating protein 2 (ENSG00000149182), score: -0.52 ARFGAP3ADP-ribosylation factor GTPase activating protein 3 (ENSG00000242247), score: -0.44 ARFRP1ADP-ribosylation factor related protein 1 (ENSG00000101246), score: -0.52 ARPC5Lactin related protein 2/3 complex, subunit 5-like (ENSG00000136950), score: -0.65 ARV1ARV1 homolog (S. cerevisiae) (ENSG00000173409), score: -0.51 ATP13A5ATPase type 13A5 (ENSG00000187527), score: 0.6 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.5 ATP7AATPase, Cu++ transporting, alpha polypeptide (ENSG00000165240), score: 0.55 AURKAIP1aurora kinase A interacting protein 1 (ENSG00000175756), score: -0.46 BAHCC1BAH domain and coiled-coil containing 1 (ENSG00000171282), score: 0.61 BAT2L2HLA-B associated transcript 2-like 2 (ENSG00000117523), score: 0.59 BEST3bestrophin 3 (ENSG00000127325), score: 0.56 BET3LBET3 like (S. cerevisiae) (ENSG00000173626), score: 0.76 BLKB lymphoid tyrosine kinase (ENSG00000136573), score: 0.74 BPIL3bactericidal/permeability-increasing protein-like 3 (ENSG00000167104), score: 0.59 BRS3bombesin-like receptor 3 (ENSG00000102239), score: 0.59 BRWD3bromodomain and WD repeat domain containing 3 (ENSG00000165288), score: 0.59 C11orf74chromosome 11 open reading frame 74 (ENSG00000166352), score: -0.48 C13orf39chromosome 13 open reading frame 39 (ENSG00000139780), score: 0.75 C14orf179chromosome 14 open reading frame 179 (ENSG00000119650), score: -0.46 C16orf42chromosome 16 open reading frame 42 (ENSG00000007520), score: -0.56 C18orf22chromosome 18 open reading frame 22 (ENSG00000101546), score: -0.72 C1orf161chromosome 1 open reading frame 161 (ENSG00000173212), score: 0.47 C1orf43chromosome 1 open reading frame 43 (ENSG00000143612), score: -0.58 C1orf55chromosome 1 open reading frame 55 (ENSG00000143751), score: 0.51 C21orf29chromosome 21 open reading frame 29 (ENSG00000175894), score: 0.56 C22orf9chromosome 22 open reading frame 9 (ENSG00000100364), score: -0.47 C3orf17chromosome 3 open reading frame 17 (ENSG00000163608), score: -0.64 C5orf24chromosome 5 open reading frame 24 (ENSG00000181904), score: 0.6 C7orf16chromosome 7 open reading frame 16 (ENSG00000106341), score: 0.49 CACHD1cache domain containing 1 (ENSG00000158966), score: 0.53 CALCRLcalcitonin receptor-like (ENSG00000064989), score: 0.5 CARD9caspase recruitment domain family, member 9 (ENSG00000187796), score: 0.51 CCDC28Bcoiled-coil domain containing 28B (ENSG00000160050), score: -0.46 CCDC45coiled-coil domain containing 45 (ENSG00000141325), score: -0.57 CCDC47coiled-coil domain containing 47 (ENSG00000108588), score: -0.47 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.51 CDR2cerebellar degeneration-related protein 2, 62kDa (ENSG00000140743), score: -0.53 CHCHD8coiled-coil-helix-coiled-coil-helix domain containing 8 (ENSG00000181924), score: -0.52 CHD6chromodomain helicase DNA binding protein 6 (ENSG00000124177), score: 0.5 CHRNA5cholinergic receptor, nicotinic, alpha 5 (ENSG00000169684), score: 0.63 CIB3calcium and integrin binding family member 3 (ENSG00000141977), score: 0.65 CINPcyclin-dependent kinase 2 interacting protein (ENSG00000100865), score: -0.51 CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.56 CNGB1cyclic nucleotide gated channel beta 1 (ENSG00000070729), score: 0.55 COBRA1cofactor of BRCA1 (ENSG00000188986), score: -0.59 CPSF4cleavage and polyadenylation specific factor 4, 30kDa (ENSG00000160917), score: -0.46 CTTNBP2NLCTTNBP2 N-terminal like (ENSG00000143079), score: 0.61 CUL4Acullin 4A (ENSG00000139842), score: -0.57 CUL5cullin 5 (ENSG00000166266), score: 0.51 DBR1debranching enzyme homolog 1 (S. cerevisiae) (ENSG00000138231), score: 0.56 DDX24DEAD (Asp-Glu-Ala-Asp) box polypeptide 24 (ENSG00000089737), score: -0.51 DDX31DEAD (Asp-Glu-Ala-Asp) box polypeptide 31 (ENSG00000125485), score: 0.51 DFFBDNA fragmentation factor, 40kDa, beta polypeptide (caspase-activated DNase) (ENSG00000169598), score: 0.64 DHX32DEAH (Asp-Glu-Ala-His) box polypeptide 32 (ENSG00000089876), score: -0.71 DMAP1DNA methyltransferase 1 associated protein 1 (ENSG00000178028), score: -0.46 DNAJC9DnaJ (Hsp40) homolog, subfamily C, member 9 (ENSG00000213551), score: -0.49 DNMT3ADNA (cytosine-5-)-methyltransferase 3 alpha (ENSG00000119772), score: 0.65 DOCK11dedicator of cytokinesis 11 (ENSG00000147251), score: 0.73 DOLPP1dolichyl pyrophosphate phosphatase 1 (ENSG00000167130), score: -0.45 ELP2elongation protein 2 homolog (S. cerevisiae) (ENSG00000134759), score: -0.49 ELP4elongation protein 4 homolog (S. cerevisiae) (ENSG00000109911), score: -0.46 EN1engrailed homeobox 1 (ENSG00000163064), score: 0.58 EPGNepithelial mitogen homolog (mouse) (ENSG00000182585), score: 0.59 EPHA3EPH receptor A3 (ENSG00000044524), score: 0.59 EPHA5EPH receptor A5 (ENSG00000145242), score: 0.49 EPHX1epoxide hydrolase 1, microsomal (xenobiotic) (ENSG00000143819), score: -0.46 ERAL1Era G-protein-like 1 (E. coli) (ENSG00000132591), score: -0.46 ERGIC3ERGIC and golgi 3 (ENSG00000125991), score: -0.53 EXOSC7exosome component 7 (ENSG00000075914), score: -0.48 FAM105Bfamily with sequence similarity 105, member B (ENSG00000154124), score: -0.64 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.47 FAM120Bfamily with sequence similarity 120B (ENSG00000112584), score: -0.51 FAM122Bfamily with sequence similarity 122B (ENSG00000156504), score: 0.59 FAM166Afamily with sequence similarity 166, member A (ENSG00000188163), score: -0.63 FAM43Afamily with sequence similarity 43, member A (ENSG00000185112), score: 0.49 FANCAFanconi anemia, complementation group A (ENSG00000187741), score: -0.56 FBXL18F-box and leucine-rich repeat protein 18 (ENSG00000155034), score: 0.71 FER1L5fer-1-like 5 (C. elegans) (ENSG00000214272), score: 0.52 FIP1L1FIP1 like 1 (S. cerevisiae) (ENSG00000145216), score: -0.59 FNBP4formin binding protein 4 (ENSG00000109920), score: 0.59 FOXL2forkhead box L2 (ENSG00000183770), score: 0.66 GAAglucosidase, alpha; acid (ENSG00000171298), score: -0.49 GABREgamma-aminobutyric acid (GABA) A receptor, epsilon (ENSG00000102287), score: 0.49 GABRR2gamma-aminobutyric acid (GABA) receptor, rho 2 (ENSG00000111886), score: 0.77 GAMTguanidinoacetate N-methyltransferase (ENSG00000130005), score: -0.5 GATMglycine amidinotransferase (L-arginine:glycine amidinotransferase) (ENSG00000171766), score: -0.64 GBA2glucosidase, beta (bile acid) 2 (ENSG00000070610), score: -0.48 GFRALGDNF family receptor alpha like (ENSG00000187871), score: 0.64 GLRA4glycine receptor, alpha 4 (ENSG00000188828), score: 0.68 GORASP1golgi reassembly stacking protein 1, 65kDa (ENSG00000114745), score: -0.55 GPR139G protein-coupled receptor 139 (ENSG00000180269), score: 0.67 GRIK1glutamate receptor, ionotropic, kainate 1 (ENSG00000171189), score: 0.63 GRIN3Aglutamate receptor, ionotropic, N-methyl-D-aspartate 3A (ENSG00000198785), score: 0.5 GRPEL2GrpE-like 2, mitochondrial (E. coli) (ENSG00000164284), score: 0.52 HELBhelicase (DNA) B (ENSG00000127311), score: 0.49 HELZhelicase with zinc finger (ENSG00000198265), score: 0.58 HNRNPA0heterogeneous nuclear ribonucleoprotein A0 (ENSG00000177733), score: -0.7 HSF4heat shock transcription factor 4 (ENSG00000102878), score: -0.45 IL1RAPL2interleukin 1 receptor accessory protein-like 2 (ENSG00000189108), score: 0.56 IMPG2interphotoreceptor matrix proteoglycan 2 (ENSG00000081148), score: 0.65 INO80DINO80 complex subunit D (ENSG00000114933), score: 0.5 INPPL1inositol polyphosphate phosphatase-like 1 (ENSG00000165458), score: -0.76 KATNA1katanin p60 (ATPase-containing) subunit A 1 (ENSG00000186625), score: -0.5 KCNA10potassium voltage-gated channel, shaker-related subfamily, member 10 (ENSG00000143105), score: 0.89 KERAkeratocan (ENSG00000139330), score: 0.91 KIAA0368KIAA0368 (ENSG00000136813), score: -0.57 KIAA2022KIAA2022 (ENSG00000050030), score: 0.49 KLC4kinesin light chain 4 (ENSG00000137171), score: 0.57 KLHDC3kelch domain containing 3 (ENSG00000124702), score: -0.5 KLHL11kelch-like 11 (Drosophila) (ENSG00000178502), score: 0.5 KLHL28kelch-like 28 (Drosophila) (ENSG00000179454), score: 0.52 KREMEN1kringle containing transmembrane protein 1 (ENSG00000183762), score: -0.45 LAMP3lysosomal-associated membrane protein 3 (ENSG00000078081), score: 0.53 LOC100290337similar to damage-specific DNA binding protein 1 (ENSG00000167986), score: -0.52 LOC100291726similar to family with sequence similarity 70, member A (ENSG00000125355), score: 0.6 LONRF3LON peptidase N-terminal domain and ring finger 3 (ENSG00000175556), score: 0.48 LRP12low density lipoprotein receptor-related protein 12 (ENSG00000147650), score: 0.49 LRRC41leucine rich repeat containing 41 (ENSG00000132128), score: -0.59 LSM4LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) (ENSG00000130520), score: -0.63 MAD2L1BPMAD2L1 binding protein (ENSG00000124688), score: -0.54 MAN2C1mannosidase, alpha, class 2C, member 1 (ENSG00000140400), score: -0.56 MAOBmonoamine oxidase B (ENSG00000069535), score: -0.45 MAPKAP1mitogen-activated protein kinase associated protein 1 (ENSG00000119487), score: -0.5 MBNL3muscleblind-like 3 (Drosophila) (ENSG00000076770), score: 0.53 MCF2MCF.2 cell line derived transforming sequence (ENSG00000101977), score: 0.59 MDGA2MAM domain containing glycosylphosphatidylinositol anchor 2 (ENSG00000139915), score: 0.57 MED8mediator complex subunit 8 (ENSG00000159479), score: -0.47 METTL9methyltransferase like 9 (ENSG00000197006), score: -0.52 MFSD11major facilitator superfamily domain containing 11 (ENSG00000092931), score: -0.45 MID2midline 2 (ENSG00000080561), score: 0.51 MRPL51mitochondrial ribosomal protein L51 (ENSG00000111639), score: -0.46 MSL1male-specific lethal 1 homolog (Drosophila) (ENSG00000188895), score: 0.54 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.58 MTERFD3MTERF domain containing 3 (ENSG00000120832), score: -0.6 MTMR1myotubularin related protein 1 (ENSG00000063601), score: 0.52 MUL1mitochondrial E3 ubiquitin ligase 1 (ENSG00000090432), score: -0.52 MUTYHmutY homolog (E. coli) (ENSG00000132781), score: 0.72 MYBBP1AMYB binding protein (P160) 1a (ENSG00000132382), score: -0.48 MYOGmyogenin (myogenic factor 4) (ENSG00000122180), score: 1 NADKNAD kinase (ENSG00000008130), score: -0.47 NAT10N-acetyltransferase 10 (GCN5-related) (ENSG00000135372), score: -0.66 NFAT5nuclear factor of activated T-cells 5, tonicity-responsive (ENSG00000102908), score: 0.62 NOL6nucleolar protein family 6 (RNA-associated) (ENSG00000165271), score: -0.48 NPY2Rneuropeptide Y receptor Y2 (ENSG00000185149), score: 0.58 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: -0.45 NSMCE1non-SMC element 1 homolog (S. cerevisiae) (ENSG00000169189), score: -0.48 NUSAP1nucleolar and spindle associated protein 1 (ENSG00000137804), score: 0.75 OGG18-oxoguanine DNA glycosylase (ENSG00000114026), score: -0.53 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.58 OLIG3oligodendrocyte transcription factor 3 (ENSG00000177468), score: 0.67 ORC5Lorigin recognition complex, subunit 5-like (yeast) (ENSG00000164815), score: -0.52 OTOAotoancorin (ENSG00000155719), score: 0.62 OTOSotospiralin (ENSG00000178602), score: 0.59 P2RY1purinergic receptor P2Y, G-protein coupled, 1 (ENSG00000169860), score: 0.65 PANK3pantothenate kinase 3 (ENSG00000120137), score: 0.5 PANX3pannexin 3 (ENSG00000154143), score: 0.78 PAPOLGpoly(A) polymerase gamma (ENSG00000115421), score: -0.76 PARNpoly(A)-specific ribonuclease (deadenylation nuclease) (ENSG00000140694), score: -0.51 PDCLphosducin-like (ENSG00000136940), score: 0.6 PDE12phosphodiesterase 12 (ENSG00000174840), score: -0.46 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.49 PHACTR4phosphatase and actin regulator 4 (ENSG00000204138), score: 0.52 PHF15PHD finger protein 15 (ENSG00000043143), score: 0.56 PHLDA3pleckstrin homology-like domain, family A, member 3 (ENSG00000174307), score: -0.51 PIGHphosphatidylinositol glycan anchor biosynthesis, class H (ENSG00000100564), score: -0.45 PIK3CGphosphoinositide-3-kinase, catalytic, gamma polypeptide (ENSG00000105851), score: 0.62 PLA2G4Aphospholipase A2, group IVA (cytosolic, calcium-dependent) (ENSG00000116711), score: 0.76 PLEKHO2pleckstrin homology domain containing, family O member 2 (ENSG00000241839), score: -0.47 POF1Bpremature ovarian failure, 1B (ENSG00000124429), score: 0.55 POLE2polymerase (DNA directed), epsilon 2 (p59 subunit) (ENSG00000100479), score: 0.65 POU1F1POU class 1 homeobox 1 (ENSG00000064835), score: 0.76 PPYR1pancreatic polypeptide receptor 1 (ENSG00000204174), score: 0.59 PRDM10PR domain containing 10 (ENSG00000170325), score: 0.57 PRKAG3protein kinase, AMP-activated, gamma 3 non-catalytic subunit (ENSG00000115592), score: 0.7 PRKG2protein kinase, cGMP-dependent, type II (ENSG00000138669), score: 0.59 PSEN1presenilin 1 (ENSG00000080815), score: -0.5 PTCHD1patched domain containing 1 (ENSG00000165186), score: 0.5 QDPRquinoid dihydropteridine reductase (ENSG00000151552), score: -0.6 RAB30RAB30, member RAS oncogene family (ENSG00000137502), score: 0.49 RAD52RAD52 homolog (S. cerevisiae) (ENSG00000002016), score: -0.5 RAD54L2RAD54-like 2 (S. cerevisiae) (ENSG00000164080), score: 0.56 RAPGEF6Rap guanine nucleotide exchange factor (GEF) 6 (ENSG00000158987), score: 0.55 RBP1retinol binding protein 1, cellular (ENSG00000114115), score: -0.54 RC3H2ring finger and CCCH-type zinc finger domains 2 (ENSG00000056586), score: 0.56 RELv-rel reticuloendotheliosis viral oncogene homolog (avian) (ENSG00000162924), score: 0.56 RGS18regulator of G-protein signaling 18 (ENSG00000150681), score: 0.61 RHOrhodopsin (ENSG00000163914), score: 0.58 SAMD12sterile alpha motif domain containing 12 (ENSG00000177570), score: 0.48 SDF4stromal cell derived factor 4 (ENSG00000078808), score: -0.57 SDK2sidekick homolog 2 (chicken) (ENSG00000069188), score: 0.62 SEMA3Asema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A (ENSG00000075213), score: 0.48 SEMA3Dsema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3D (ENSG00000153993), score: 0.53 SEP1515 kDa selenoprotein (ENSG00000183291), score: -0.59 SEPT2septin 2 (ENSG00000168385), score: -0.52 SERTAD4SERTA domain containing 4 (ENSG00000082497), score: 0.52 SESTD1SEC14 and spectrin domains 1 (ENSG00000187231), score: 0.52 SGCZsarcoglycan, zeta (ENSG00000185053), score: 0.5 SIKE1suppressor of IKBKE 1 (ENSG00000052723), score: 0.5 SKIV2Lsuperkiller viralicidic activity 2-like (S. cerevisiae) (ENSG00000204351), score: -0.45 SLBPstem-loop binding protein (ENSG00000163950), score: -0.88 SLC25A37solute carrier family 25, member 37 (ENSG00000147454), score: -0.49 SLC26A5solute carrier family 26, member 5 (prestin) (ENSG00000170615), score: 0.59 SLC9A8solute carrier family 9 (sodium/hydrogen exchanger), member 8 (ENSG00000197818), score: 0.54 SNX3sorting nexin 3 (ENSG00000112335), score: -0.58 SPP1secreted phosphoprotein 1 (ENSG00000118785), score: -0.46 SPPL2Bsignal peptide peptidase-like 2B (ENSG00000005206), score: -0.48 SPRED1sprouty-related, EVH1 domain containing 1 (ENSG00000166068), score: 0.48 SRRM1serine/arginine repetitive matrix 1 (ENSG00000133226), score: 0.57 SSNA1Sjogren syndrome nuclear autoantigen 1 (ENSG00000176101), score: -0.66 ST8SIA2ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ENSG00000140557), score: 0.55 STK11serine/threonine kinase 11 (ENSG00000118046), score: -0.46 STXBP4syntaxin binding protein 4 (ENSG00000166263), score: 0.59 SURF1surfeit 1 (ENSG00000148290), score: -0.5 SYPL1synaptophysin-like 1 (ENSG00000008282), score: -0.79 TT, brachyury homolog (mouse) (ENSG00000164458), score: 0.8 TAB3TGF-beta activated kinase 1/MAP3K7 binding protein 3 (ENSG00000157625), score: 0.51 TADA2Btranscriptional adaptor 2B (ENSG00000173011), score: -0.45 TAF15TAF15 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 68kDa (ENSG00000172660), score: -0.47 TAF3TAF3 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 140kDa (ENSG00000165632), score: 0.58 TAF5TAF5 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 100kDa (ENSG00000148835), score: 0.71 TAOK1TAO kinase 1 (ENSG00000160551), score: 0.53 TBX18T-box 18 (ENSG00000112837), score: 0.52 TCF12transcription factor 12 (ENSG00000140262), score: 0.59 TIRAPtoll-interleukin 1 receptor (TIR) domain containing adaptor protein (ENSG00000150455), score: 0.5 TLL1tolloid-like 1 (ENSG00000038295), score: 0.64 TM4SF1transmembrane 4 L six family member 1 (ENSG00000169908), score: -0.55 TMEM128transmembrane protein 128 (ENSG00000132406), score: -0.45 TMEM22transmembrane protein 22 (ENSG00000168917), score: 0.52 TMEM41Atransmembrane protein 41A (ENSG00000163900), score: 0.51 TMEM43transmembrane protein 43 (ENSG00000170876), score: -0.45 TMTC3transmembrane and tetratricopeptide repeat containing 3 (ENSG00000139324), score: 0.51 TMX1thioredoxin-related transmembrane protein 1 (ENSG00000139921), score: -0.71 TNIKTRAF2 and NCK interacting kinase (ENSG00000154310), score: 0.54 TOR2Atorsin family 2, member A (ENSG00000160404), score: -0.56 TRABDTraB domain containing (ENSG00000170638), score: -0.61 TSTA3tissue specific transplantation antigen P35B (ENSG00000104522), score: -0.51 TUBB1tubulin, beta 1 (ENSG00000101162), score: 0.53 TXNRD1thioredoxin reductase 1 (ENSG00000198431), score: -0.47 UBAP1ubiquitin associated protein 1 (ENSG00000165006), score: -0.73 UBE2Zubiquitin-conjugating enzyme E2Z (ENSG00000159202), score: -0.56 USP45ubiquitin specific peptidase 45 (ENSG00000123552), score: 0.52 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.52 VTI1Avesicle transport through interaction with t-SNAREs homolog 1A (yeast) (ENSG00000151532), score: 0.51 VWA5B1von Willebrand factor A domain containing 5B1 (ENSG00000158816), score: 0.48 WDR34WD repeat domain 34 (ENSG00000119333), score: -0.52 WHSC1L1Wolf-Hirschhorn syndrome candidate 1-like 1 (ENSG00000147548), score: 0.51 WRNIP1Werner helicase interacting protein 1 (ENSG00000124535), score: -0.6 ZBTB33zinc finger and BTB domain containing 33 (ENSG00000177485), score: 0.5 ZBTB37zinc finger and BTB domain containing 37 (ENSG00000185278), score: 0.65 ZBTB40zinc finger and BTB domain containing 40 (ENSG00000184677), score: -0.56 ZBTB8Bzinc finger and BTB domain containing 8B (ENSG00000215897), score: 0.66 ZC3H12Bzinc finger CCCH-type containing 12B (ENSG00000102053), score: 0.57 ZCCHC4zinc finger, CCHC domain containing 4 (ENSG00000168228), score: 0.55 ZNF828zinc finger protein 828 (ENSG00000198824), score: 0.59
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ht_f_ca1 | gga | ht | f | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| gga_br_m_ca1 | gga | br | m | _ |
| gga_br_f_ca1 | gga | br | f | _ |
| gga_cb_m_ca1 | gga | cb | m | _ |
| gga_cb_f_ca1 | gga | cb | f | _ |