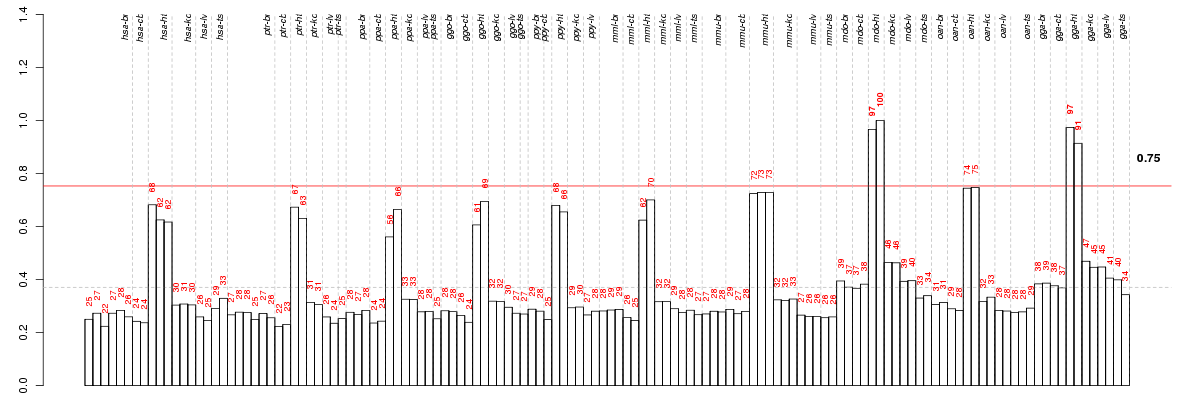

Under-expression is coded with green,
over-expression with red color.

metabolic process
The chemical reactions and pathways, including anabolism and catabolism, by which living organisms transform chemical substances. Metabolic processes typically transform small molecules, but also include macromolecular processes such as DNA repair and replication, and protein synthesis and degradation.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
system process
A multicellular organismal process carried out by any of the organs or tissues in an organ system. An organ system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a biological objective.
muscle system process
A organ system process carried out at the level of a muscle. Muscle tissue is composed of contractile cells or fibers.
circulatory system process
A organ system process carried out by any of the organs or tissues of the circulatory system. The circulatory system is an organ system that moves extracellular fluids to and from tissue within a multicellular organism.
heart process
A circulatory system process carried out by the heart. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
muscle contraction
A process whereby force is generated within muscle tissue, resulting in a change in muscle geometry. Force generation involves a chemo-mechanical energy conversion step that is carried out by the actin/myosin complex activity, which generates force through ATP hydrolysis.
generation of precursor metabolites and energy
The chemical reactions and pathways resulting in the formation of precursor metabolites, substances from which energy is derived, and any process involved in the liberation of energy from these substances.
oxidation reduction
The process of removal or addition of one or more electrons with or without the concomitant removal or addition of a proton or protons.
respiratory electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors such as NADH and FADH2 to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
oxidative phosphorylation
The phosphorylation of ADP to ATP that accompanies the oxidation of a metabolite through the operation of the respiratory chain. Oxidation of compounds establishes a proton gradient across the membrane, providing the energy for ATP synthesis.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
phosphorus metabolic process
The chemical reactions and pathways involving the nonmetallic element phosphorus or compounds that contain phosphorus, usually in the form of a phosphate group (PO4).
phosphate metabolic process
The chemical reactions and pathways involving the phosphate group, the anion or salt of any phosphoric acid.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
heart development
The process whose specific outcome is the progression of the heart over time, from its formation to the mature structure. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
blood circulation
The flow of blood through the body of an animal, enabling the transport of nutrients to the tissues and the removal of waste products.
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
biological_process
Any process specifically pertinent to the functioning of integrated living units: cells, tissues, organs, and organisms. A process is a collection of molecular events with a defined beginning and end.
aerobic respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which requires oxygen as the terminal electron acceptor.
cellular process
Any process that is carried out at the cellular level, but not necessarily restricted to a single cell. For example, cell communication occurs among more than one cell, but occurs at the cellular level.
striated muscle tissue development
The process whose specific outcome is the progression of a striated muscle over time, from its formation to the mature structure. Striated muscle contain fibers that are divided by transverse bands into striations, and cardiac and skeletal muscle are types of striated muscle. Skeletal muscle myoblasts fuse to form myotubes and eventually multinucleated muscle fibers. The fusion of cardiac cells is very rare and can only form binucleate cells.
energy derivation by oxidation of organic compounds
The chemical reactions and pathways by which a cell derives energy from organic compounds; results in the oxidation of the compounds from which energy is released.
phosphorylation
The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide.
multicellular organismal process
Any biological process, occurring at the level of a multicellular organism, pertinent to its function.
developmental process
A biological process whose specific outcome is the progression of an integrated living unit: an anatomical structure (which may be a subcellular structure, cell, tissue, or organ), or organism over time from an initial condition to a later condition.
ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP.
mitochondrial ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP, as it occurs in the mitochondrial inner membrane or chloroplast thylakoid membrane.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
cellular respiration
The enzymatic release of energy from organic compounds (especially carbohydrates and fats) which either requires oxygen (aerobic respiration) or does not (anaerobic respiration).
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
anatomical structure development
The biological process whose specific outcome is the progression of an anatomical structure from an initial condition to its mature state. This process begins with the formation of the structure and ends with the mature structure, whatever form that may be including its natural destruction. An anatomical structure is any biological entity that occupies space and is distinguished from its surroundings. Anatomical structures can be macroscopic such as a carpel, or microscopic such as an acrosome.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
muscle tissue morphogenesis
The process by which the anatomical structures of muscle tissue are generated and organized. Muscle tissue consists of a set of cells that are part of an organ and carry out a contractive function. Morphogenesis pertains to the creation of form.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
muscle structure development
The progression of a muscle structure over time, from its formation to its mature state. Muscle structures are contractile cells, tissues or organs that are found in multicellular organisms.
biological regulation
Any process that modulates the frequency, rate or extent of any biological process, quality or function.
regulation of biological quality
Any process that modulates the frequency, rate or extent of a biological quality. A biological quality is a measurable attribute of an organism or part of an organism, such as size, mass, shape, color, etc.
all
NA
cellular metabolic process
The chemical reactions and pathways by which individual cells transform chemical substances.
multicellular organismal development
The biological process whose specific outcome is the progression of a multicellular organism over time from an initial condition (e.g. a zygote or a young adult) to a later condition (e.g. a multicellular animal or an aged adult).
regulation of multicellular organismal process
Any process that modulates the frequency, rate or extent of a multicellular organismal process, the processes pertinent to the function of a multicellular organism above the cellular level; includes the integrated processes of tissues and organs.
regulation of biological process
Any process that modulates the frequency, rate or extent of a biological process. Biological processes are regulated by many means; examples include the control of gene expression, protein modification or interaction with a protein or substrate molecule.
regulation of system process
Any process that modulates the frequency, rate or extent of a system process, a multicellular organismal process carried out by any of the organs or tissues in an organ system.
anatomical structure morphogenesis
The process by which anatomical structures are generated and organized. Morphogenesis pertains to the creation of form.
system development
The process whose specific outcome is the progression of an organismal system over time, from its formation to the mature structure. A system is a regularly interacting or interdependent group of organs or tissues that work together to carry out a given biological process.
electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
organ development
Development of a tissue or tissues that work together to perform a specific function or functions. Development pertains to the process whose specific outcome is the progression of a structure over time, from its formation to the mature structure. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
tissue morphogenesis
The process by which the anatomical structures of a tissue are generated and organized. Morphogenesis pertains to the creation of form.
organ morphogenesis
Morphogenesis of an organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process by which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions.
tissue development
The process whose specific outcome is the progression of a tissue over time, from its formation to the mature structure.
muscle organ development
The process whose specific outcome is the progression of the muscle over time, from its formation to the mature structure. The muscle is an organ consisting of a tissue made up of various elongated cells that are specialized to contract and thus to produce movement and mechanical work.
ATP synthesis coupled electron transport
The transfer of electrons through a series of electron donors and acceptors, generating energy that is ultimately used for synthesis of ATP.
heart contraction
The multicellular organismal process by which the heart decreases in volume in a characteristic way to propel blood through the body.
regulation of the force of heart contraction
Any process that modulates the extent of heart contraction, changing the force with which blood is propelled.
cardiac chamber morphogenesis
The process by which a cardiac chamber is generated and organized. A cardiac chamber is an enclosed cavity within the heart.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.
heart morphogenesis
The developmental process by which the heart is generated and organized. The heart is a hollow, muscular organ, which, by contracting rhythmically, keeps up the circulation of the blood.
cardiac chamber development
The progression of a cardiac chamber over time, from its formation to the mature structure. A cardiac chamber is an enclosed cavity within the heart.
muscle tissue development
The progression of muscle tissue over time, from its initial formation to its mature state. Muscle tissue is a contractile tissue made up of actin and myosin fibers.
respiratory electron transport chain
A process whereby a series of electron carriers operate together to transfer electrons from donors such as NADH and FADH2 to any of several different terminal electron acceptors to generate a transmembrane electrochemical gradient.
oxidative phosphorylation
The phosphorylation of ADP to ATP that accompanies the oxidation of a metabolite through the operation of the respiratory chain. Oxidation of compounds establishes a proton gradient across the membrane, providing the energy for ATP synthesis.
regulation of heart contraction
Any process that modulates the frequency, rate or extent of heart contraction. Heart contraction is the process by which the heart decreases in volume in a characteristic way to propel blood through the body.
cardiac muscle tissue development
The process whose specific outcome is the progression of cardiac muscle over time, from its formation to the mature structure.
cardiac muscle tissue morphogenesis
The process by which the anatomical structures of cardiac muscle tissue are generated and organized. Morphogenesis pertains to the creation of form.

plasma membrane
The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
intracellular
The living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
membrane
Double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cellular_component
The part of a cell or its extracellular environment in which a gene product is located. A gene product may be located in one or more parts of a cell and its location may be as specific as a particular macromolecular complex, that is, a stable, persistent association of macromolecules that function together.
cell
The basic structural and functional unit of all organisms. Includes the plasma membrane and any external encapsulating structures such as the cell wall and cell envelope.
cytoplasm
All of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
cytoskeleton
Any of the various filamentous elements that form the internal framework of cells, and typically remain after treatment of the cells with mild detergent to remove membrane constituents and soluble components of the cytoplasm. The term embraces intermediate filaments, microfilaments, microtubules, the microtrabecular lattice, and other structures characterized by a polymeric filamentous nature and long-range order within the cell. The various elements of the cytoskeleton not only serve in the maintenance of cellular shape but also have roles in other cellular functions, including cellular movement, cell division, endocytosis, and movement of organelles.
actin cytoskeleton
The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
myofibril
The contractile element of skeletal and cardiac muscle; a long, highly organized bundle of actin, myosin, and other proteins that contracts by a sliding filament mechanism.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
T-tubule
Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle membrane
The lipid bilayer surrounding an organelle.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
envelope
A multilayered structure surrounding all or part of a cell; encompasses one or more lipid bilayers, and may include a cell wall layer; also includes the space between layers.
macromolecular complex
A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which the constituent parts function together.
sarcolemma
The outer membrane of a muscle cell, consisting of the plasma membrane, a covering basement membrane (about 100 nm thick and sometimes common to more than one fiber), and the associated loose network of collagen fibers.
organelle
Organized structure of distinctive morphology and function. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, and prokaryotic structures such as anammoxosomes and pirellulosomes. Excludes the plasma membrane.
membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
protein complex
Any macromolecular complex composed of two or more polypeptide subunits, which may or may not be identical. Protein complexes may have other associated non-protein prosthetic groups, such as nucleotides, metal ions or other small molecules.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
respiratory chain
The protein complexes that form the electron transport system (the respiratory chain), associated with a cell membrane, usually the plasma membrane (in prokaryotes) or the inner mitochondrial membrane (on eukaryotes). The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
all
NA
cell part
Any constituent part of a cell, the basic structural and functional unit of all organisms.
organelle part
Any constituent part of an organelle, an organized structure of distinctive morphology and function. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton, but excludes the plasma membrane.
organelle membrane
The lipid bilayer surrounding an organelle.
intracellular membrane-bounded organelle
Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane.
intracellular non-membrane-bounded organelle
Organized structure of distinctive morphology and function, not bounded by a lipid bilayer membrane and occurring within the cell. Includes ribosomes, the cytoskeleton and chromosomes.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
intracellular part
Any constituent part of the living contents of a cell; the matter contained within (but not including) the plasma membrane, usually taken to exclude large vacuoles and masses of secretory or ingested material. In eukaryotes it includes the nucleus and cytoplasm.
organelle membrane
The lipid bilayer surrounding an organelle.
membrane part
Any constituent part of a membrane, a double layer of lipid molecules that encloses all cells, and, in eukaryotes, many organelles; may be a single or double lipid bilayer; also includes associated proteins.
organelle envelope
A double membrane structure enclosing an organelle, including two lipid bilayers and the region between them. In some cases, an organelle envelope may have more than two membranes.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
intracellular organelle
Organized structure of distinctive morphology and function, occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton. Excludes the plasma membrane.
intracellular organelle part
A constituent part of an intracellular organelle, an organized structure of distinctive morphology and function, occurring within the cell. Includes constituent parts of the nucleus, mitochondria, plastids, vacuoles, vesicles, ribosomes and the cytoskeleton but excludes the plasma membrane.
NADH dehydrogenase complex
An integral membrane complex that possesses NADH oxidoreductase activity. The complex is one of the components of the electron transport chain. It catalyzes the transfer of a pair of electrons from NADH to a quinone.
organelle inner membrane
The inner, i.e. lumen-facing, lipid bilayer of an organelle envelope; usually highly selective to most ions and metabolites.
mitochondrial inner membrane
The inner, i.e. lumen-facing, lipid bilayer of the mitochondrial envelope. It is highly folded to form cristae.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial envelope
The double lipid bilayer enclosing the mitochondrion and separating its contents from the cell cytoplasm; includes the intermembrane space.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial membrane part
Any constituent part of a mitochondrial membrane, either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
A band
The dark-staining region of a sarcomere, in which myosin thick filaments are present; the center is traversed by the paler H zone, which in turn contains the M line.
I band
A region of a sarcomere that appears as a light band on each side of the Z disc, comprising a region of the sarcomere where thin (actin) filaments are not overlapped by thick (myosin) filaments; contains actin, troponin, and tropomyosin; each sarcomere includes half of an I band at each end.
Z disc
Platelike region of a muscle sarcomere to which the plus ends of actin filaments are attached.
plasma membrane part
Any constituent part of the plasma membrane, the membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins.
cytoplasmic part
Any constituent part of the cytoplasm, all of the contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures.
contractile fiber
Fibers, composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrion
A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
respiratory chain complex I
Respiratory chain complex I is an enzyme of the respiratory chain. It consists of at least 34 polypeptide chains and is L-shaped, with a horizontal arm lying in the membrane and a vertical arm that projects into the matrix. The electrons of NADH enter the chain at this complex.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
mitochondrial membrane
Either of the lipid bilayers that surround the mitochondrion and form the mitochondrial envelope.
mitochondrial part
Any constituent part of a mitochondrion, a semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration.
contractile fiber part
Any constituent part of a contractile fiber, a fiber composed of actin, myosin, and associated proteins, found in cells of smooth or striated muscle.
mitochondrial respiratory chain
The protein complexes that form the mitochondrial electron transport system (the respiratory chain), associated with the inner mitochondrial membrane. The respiratory chain complexes transfer electrons from an electron donor to an electron acceptor and are associated with a proton pump to create a transmembrane electrochemical gradient.
T-tubule
Invagination of the plasma membrane of a muscle cell that extends inward from the cell surface around each myofibril. The ends of T-tubules make contact with the sarcoplasmic reticulum membrane.
mitochondrial respiratory chain complex I
A protein complex located in the mitochondrial inner membrane that forms part of the mitochondrial respiratory chain. It contains about 25 different polypeptide subunits, including NADH dehydrogenase (ubiquinone), flavin mononucleotide and several different iron-sulfur clusters containing non-heme iron. The iron undergoes oxidation-reduction between Fe(II) and Fe(III), and catalyzes proton translocation linked to the oxidation of NADH by ubiquinone.
sarcomere
The repeating unit of a myofibril in a muscle cell, composed of an array of overlapping thick and thin filaments between two adjacent Z discs.

molecular_function
Elemental activities, such as catalysis or binding, describing the actions of a gene product at the molecular level. A given gene product may exhibit one or more molecular functions.
catalytic activity
Catalysis of a biochemical reaction at physiological temperatures. In biologically catalyzed reactions, the reactants are known as substrates, and the catalysts are naturally occurring macromolecular substances known as enzymes. Enzymes possess specific binding sites for substrates, and are usually composed wholly or largely of protein, but RNA that has catalytic activity (ribozyme) is often also regarded as enzymatic.
NADH dehydrogenase activity
Catalysis of the reaction: NADH + H+ + acceptor = NAD+ + reduced acceptor.
oxidoreductase activity
Catalysis of an oxidation-reduction (redox) reaction, a reversible chemical reaction in which the oxidation state of an atom or atoms within a molecule is altered. One substrate acts as a hydrogen or electron donor and becomes oxidized, while the other acts as hydrogen or electron acceptor and becomes reduced.
NADH dehydrogenase (ubiquinone) activity
Catalysis of the reaction: NADH + H+ + ubiquinone = NAD+ + ubiquinol.
oxidoreductase activity, acting on NADH or NADPH
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a hydrogen or electron acceptor.
oxidoreductase activity, acting on NADH or NADPH, quinone or similar compound as acceptor
Catalysis of an oxidation-reduction (redox) reaction in which NADH or NADPH acts as a hydrogen or electron donor and reduces a quinone or a similar acceptor molecule.
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
all
NA
NADH dehydrogenase (quinone) activity
Catalysis of the reaction: NADH + H+ + a quinone = NAD+ + a quinol.
| Id | Pvalue | ExpCount | Count | Size | Term |
|---|---|---|---|---|---|
| 04260 | 3.613e-05 | 1.327 | 10 | 25 | Cardiac muscle contraction |
| 05410 | 5.699e-04 | 1.805 | 10 | 34 | Hypertrophic cardiomyopathy (HCM) |
| 05414 | 3.153e-03 | 1.805 | 9 | 34 | Dilated cardiomyopathy |
| 05010 | 4.334e-03 | 2.335 | 10 | 44 | Alzheimer's disease |
| 00190 | 6.761e-03 | 1.592 | 8 | 30 | Oxidative phosphorylation |
| 05012 | 1.773e-02 | 1.433 | 7 | 27 | Parkinson's disease |
| 05016 | 3.146e-02 | 2.601 | 9 | 49 | Huntington's disease |
ACAD8acyl-CoA dehydrogenase family, member 8 (ENSG00000151498), score: 0.56 ACAD9acyl-CoA dehydrogenase family, member 9 (ENSG00000177646), score: 0.57 ACANaggrecan (ENSG00000157766), score: 0.72 ACOT11acyl-CoA thioesterase 11 (ENSG00000162390), score: 0.5 ACSL3acyl-CoA synthetase long-chain family member 3 (ENSG00000123983), score: -0.65 ACTN2actinin, alpha 2 (ENSG00000077522), score: 0.47 ADAMTS8ADAM metallopeptidase with thrombospondin type 1 motif, 8 (ENSG00000134917), score: 0.44 ADAMTSL5ADAMTS-like 5 (ENSG00000185761), score: 0.44 ADAT1adenosine deaminase, tRNA-specific 1 (ENSG00000065457), score: 0.51 ADSLadenylosuccinate lyase (ENSG00000100354), score: 0.42 AICDAactivation-induced cytidine deaminase (ENSG00000111732), score: 0.8 ALDH6A1aldehyde dehydrogenase 6 family, member A1 (ENSG00000119711), score: 0.42 ALKBH1alkB, alkylation repair homolog 1 (E. coli) (ENSG00000100601), score: 0.45 ANKRD1ankyrin repeat domain 1 (cardiac muscle) (ENSG00000148677), score: 0.41 ANKRD42ankyrin repeat domain 42 (ENSG00000137494), score: -0.45 APOBEC2apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 2 (ENSG00000124701), score: 0.56 ARHGAP40Rho GTPase activating protein 40 (ENSG00000124143), score: 0.46 ARHGEF6Rac/Cdc42 guanine nucleotide exchange factor (GEF) 6 (ENSG00000129675), score: 0.41 ARNTaryl hydrocarbon receptor nuclear translocator (ENSG00000143437), score: 0.52 ASB15ankyrin repeat and SOCS box-containing 15 (ENSG00000146809), score: 0.48 ASB2ankyrin repeat and SOCS box-containing 2 (ENSG00000100628), score: 0.46 ATG16L1ATG16 autophagy related 16-like 1 (S. cerevisiae) (ENSG00000085978), score: 0.54 ATP1B4ATPase, Na+/K+ transporting, beta 4 polypeptide (ENSG00000101892), score: 0.79 ATP2A2ATPase, Ca++ transporting, cardiac muscle, slow twitch 2 (ENSG00000174437), score: 0.57 ATPIF1ATPase inhibitory factor 1 (ENSG00000130770), score: 0.47 BEST3bestrophin 3 (ENSG00000127325), score: 0.81 BICD2bicaudal D homolog 2 (Drosophila) (ENSG00000185963), score: 0.44 BLVRAbiliverdin reductase A (ENSG00000106605), score: -0.6 C10orf137chromosome 10 open reading frame 137 (ENSG00000107938), score: 0.46 C12orf5chromosome 12 open reading frame 5 (ENSG00000078237), score: 0.45 C1QTNF7C1q and tumor necrosis factor related protein 7 (ENSG00000163145), score: 0.46 C3orf23chromosome 3 open reading frame 23 (ENSG00000179152), score: 0.5 C6orf106chromosome 6 open reading frame 106 (ENSG00000196821), score: -0.46 C6orf142chromosome 6 open reading frame 142 (ENSG00000146147), score: 0.56 CA13carbonic anhydrase XIII (ENSG00000185015), score: 0.55 CABC1chaperone, ABC1 activity of bc1 complex homolog (S. pombe) (ENSG00000163050), score: 0.42 CACNA1Ccalcium channel, voltage-dependent, L type, alpha 1C subunit (ENSG00000151067), score: 0.42 CADM1cell adhesion molecule 1 (ENSG00000182985), score: -0.55 CALCRLcalcitonin receptor-like (ENSG00000064989), score: 0.46 CCNYcyclin Y (ENSG00000108100), score: -0.47 CDC123cell division cycle 123 homolog (S. cerevisiae) (ENSG00000151465), score: -0.67 CDCP2CUB domain containing protein 2 (ENSG00000157211), score: 0.54 CDH2cadherin 2, type 1, N-cadherin (neuronal) (ENSG00000170558), score: 0.42 CDK6cyclin-dependent kinase 6 (ENSG00000105810), score: 0.43 CHMP7CHMP family, member 7 (ENSG00000147457), score: 0.49 CHUKconserved helix-loop-helix ubiquitous kinase (ENSG00000213341), score: 0.43 CLEC3AC-type lectin domain family 3, member A (ENSG00000166509), score: 0.57 CLIC4chloride intracellular channel 4 (ENSG00000169504), score: 0.41 CLMNcalmin (calponin-like, transmembrane) (ENSG00000165959), score: -0.46 CLN6ceroid-lipofuscinosis, neuronal 6, late infantile, variant (ENSG00000128973), score: 0.47 CMAScytidine monophosphate N-acetylneuraminic acid synthetase (ENSG00000111726), score: -0.52 CMYA5cardiomyopathy associated 5 (ENSG00000164309), score: 0.5 COL4A1collagen, type IV, alpha 1 (ENSG00000187498), score: 0.45 COQ9coenzyme Q9 homolog (S. cerevisiae) (ENSG00000088682), score: 0.51 COX10COX10 homolog, cytochrome c oxidase assembly protein, heme A: farnesyltransferase (yeast) (ENSG00000006695), score: 0.44 CRYGNcrystallin, gamma N (ENSG00000127377), score: 0.53 CSRP3cysteine and glycine-rich protein 3 (cardiac LIM protein) (ENSG00000129170), score: 0.52 CTTNBP2NLCTTNBP2 N-terminal like (ENSG00000143079), score: 0.57 DACH1dachshund homolog 1 (Drosophila) (ENSG00000165659), score: 0.51 DAPP1dual adaptor of phosphotyrosine and 3-phosphoinositides (ENSG00000070190), score: 0.42 DCBLD1discoidin, CUB and LCCL domain containing 1 (ENSG00000164465), score: 0.43 DDX47DEAD (Asp-Glu-Ala-Asp) box polypeptide 47 (ENSG00000213782), score: -0.55 DENND5BDENN/MADD domain containing 5B (ENSG00000170456), score: 0.48 DHX35DEAH (Asp-Glu-Ala-His) box polypeptide 35 (ENSG00000101452), score: -0.46 DLSTdihydrolipoamide S-succinyltransferase (E2 component of 2-oxo-glutarate complex) (ENSG00000119689), score: 0.42 DOLPP1dolichyl pyrophosphate phosphatase 1 (ENSG00000167130), score: -0.49 DUSP27dual specificity phosphatase 27 (putative) (ENSG00000198842), score: 0.44 EDNRAendothelin receptor type A (ENSG00000151617), score: 0.73 EGLN1egl nine homolog 1 (C. elegans) (ENSG00000135766), score: 0.56 EIF3Deukaryotic translation initiation factor 3, subunit D (ENSG00000100353), score: 0.49 EIF4EBP1eukaryotic translation initiation factor 4E binding protein 1 (ENSG00000187840), score: 0.42 ERICH1glutamate-rich 1 (ENSG00000104714), score: -0.46 EXD2exonuclease 3'-5' domain containing 2 (ENSG00000081177), score: -0.49 F2RL2coagulation factor II (thrombin) receptor-like 2 (ENSG00000164220), score: 0.63 FAM101Bfamily with sequence similarity 101, member B (ENSG00000183688), score: 0.47 FAM109Bfamily with sequence similarity 109, member B (ENSG00000177096), score: 0.45 FAM40Bfamily with sequence similarity 40, member B (ENSG00000128578), score: 0.45 FAM54Bfamily with sequence similarity 54, member B (ENSG00000117640), score: 0.47 FBP2fructose-1,6-bisphosphatase 2 (ENSG00000130957), score: 0.75 FBXO3F-box protein 3 (ENSG00000110429), score: 0.49 FBXO32F-box protein 32 (ENSG00000156804), score: 0.45 FBXO40F-box protein 40 (ENSG00000163833), score: 0.45 FER1L5fer-1-like 5 (C. elegans) (ENSG00000214272), score: 0.45 FGF4fibroblast growth factor 4 (ENSG00000075388), score: 0.56 FGFR3fibroblast growth factor receptor 3 (ENSG00000068078), score: -0.48 FHL2four and a half LIM domains 2 (ENSG00000115641), score: 0.55 FHL3four and a half LIM domains 3 (ENSG00000183386), score: 0.7 FILIP1filamin A interacting protein 1 (ENSG00000118407), score: 0.55 FSD2fibronectin type III and SPRY domain containing 2 (ENSG00000186628), score: 0.58 FZD2frizzled homolog 2 (Drosophila) (ENSG00000180340), score: 0.43 GATA5GATA binding protein 5 (ENSG00000130700), score: 0.83 GATA6GATA binding protein 6 (ENSG00000141448), score: 0.42 GBGT1globoside alpha-1,3-N-acetylgalactosaminyltransferase 1 (ENSG00000148288), score: 0.44 GCNT7glucosaminyl (N-acetyl) transferase family member 7 (ENSG00000124091), score: 0.45 GJA5gap junction protein, alpha 5, 40kDa (ENSG00000143140), score: 0.43 GJD4gap junction protein, delta 4, 40.1kDa (ENSG00000177291), score: 0.46 GOT1glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) (ENSG00000120053), score: 0.43 GPR107G protein-coupled receptor 107 (ENSG00000148358), score: -0.54 GPT2glutamic pyruvate transaminase (alanine aminotransferase) 2 (ENSG00000166123), score: -0.53 HCCSholocytochrome c synthase (ENSG00000004961), score: 0.48 HMG20Ahigh-mobility group 20A (ENSG00000140382), score: -0.48 HMGXB4HMG box domain containing 4 (ENSG00000100281), score: 0.42 HSPG2heparan sulfate proteoglycan 2 (ENSG00000142798), score: 0.51 IDH3Aisocitrate dehydrogenase 3 (NAD+) alpha (ENSG00000166411), score: 0.43 IFT27intraflagellar transport 27 homolog (Chlamydomonas) (ENSG00000100360), score: -0.49 IFT57intraflagellar transport 57 homolog (Chlamydomonas) (ENSG00000114446), score: -0.45 IL15interleukin 15 (ENSG00000164136), score: 0.5 KBTBD10kelch repeat and BTB (POZ) domain containing 10 (ENSG00000239474), score: 0.45 KCNJ2potassium inwardly-rectifying channel, subfamily J, member 2 (ENSG00000123700), score: 0.49 KHDRBS3KH domain containing, RNA binding, signal transduction associated 3 (ENSG00000131773), score: -0.46 KIAA0226KIAA0226 (ENSG00000145016), score: 0.5 KITLGKIT ligand (ENSG00000049130), score: 0.41 KLF8Kruppel-like factor 8 (ENSG00000102349), score: 0.43 KLHDC2kelch domain containing 2 (ENSG00000165516), score: 0.42 KLHL24kelch-like 24 (Drosophila) (ENSG00000114796), score: 0.46 KLHL31kelch-like 31 (Drosophila) (ENSG00000124743), score: 0.65 KLHL38kelch-like 38 (Drosophila) (ENSG00000175946), score: 0.43 LACE1lactation elevated 1 (ENSG00000135537), score: 0.43 LASS6LAG1 homolog, ceramide synthase 6 (ENSG00000172292), score: -0.52 LBHlimb bud and heart development homolog (mouse) (ENSG00000213626), score: 0.44 LDB3LIM domain binding 3 (ENSG00000122367), score: 0.53 LMOD2leiomodin 2 (cardiac) (ENSG00000170807), score: 0.53 LOC100131509similar to inward rectifying K+ channel negative regulator Kir2.2v (ENSG00000184185), score: 0.61 LOC100134291similar to mitogen-activated protein kinase phosphatase x (ENSG00000112679), score: 0.61 LOC100291405similar to protein tyrosine phosphatase-like A domain containing 1 (ENSG00000074696), score: -0.45 LOC100291671similar to SH3-binding domain and glutamic acid-rich protein (ENSG00000185437), score: 0.48 LPHN2latrophilin 2 (ENSG00000117114), score: 0.42 LRRC10leucine rich repeat containing 10 (ENSG00000198812), score: 0.67 LRRC20leucine rich repeat containing 20 (ENSG00000172731), score: 0.47 LSSlanosterol synthase (2,3-oxidosqualene-lanosterol cyclase) (ENSG00000160285), score: -0.48 LYPD6LY6/PLAUR domain containing 6 (ENSG00000187123), score: 0.54 MAP7microtubule-associated protein 7 (ENSG00000135525), score: -0.6 MAPKSP1MAPK scaffold protein 1 (ENSG00000109270), score: 0.48 MEOX2mesenchyme homeobox 2 (ENSG00000106511), score: 0.55 MESDC1mesoderm development candidate 1 (ENSG00000140406), score: 0.49 MFN1mitofusin 1 (ENSG00000171109), score: 0.64 MFSD10major facilitator superfamily domain containing 10 (ENSG00000109736), score: -0.47 MRPS14mitochondrial ribosomal protein S14 (ENSG00000120333), score: 0.46 MST4serine/threonine protein kinase MST4 (ENSG00000134602), score: 0.51 MTERFD1MTERF domain containing 1 (ENSG00000156469), score: 0.55 MTMR1myotubularin related protein 1 (ENSG00000063601), score: 0.47 MYBPC3myosin binding protein C, cardiac (ENSG00000134571), score: 0.55 MYCT1myc target 1 (ENSG00000120279), score: 0.48 MYL3myosin, light chain 3, alkali; ventricular, skeletal, slow (ENSG00000160808), score: 0.52 MYLK3myosin light chain kinase 3 (ENSG00000140795), score: 0.56 MYOM2myomesin (M-protein) 2, 165kDa (ENSG00000036448), score: 0.43 MYOTmyotilin (ENSG00000120729), score: 1 MYOZ2myozenin 2 (ENSG00000172399), score: 0.53 MYPNmyopalladin (ENSG00000138347), score: 0.42 NDUFA10NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 10, 42kDa (ENSG00000130414), score: 0.57 NDUFA12NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 12 (ENSG00000184752), score: 0.61 NDUFA6NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 6, 14kDa (ENSG00000184983), score: 0.47 NDUFB10NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 10, 22kDa (ENSG00000140990), score: 0.52 NDUFS3NADH dehydrogenase (ubiquinone) Fe-S protein 3, 30kDa (NADH-coenzyme Q reductase) (ENSG00000213619), score: 0.47 NFATC1nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 1 (ENSG00000131196), score: 0.41 NFU1NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) (ENSG00000169599), score: 0.48 NHSNance-Horan syndrome (congenital cataracts and dental anomalies) (ENSG00000188158), score: 0.5 NIPSNAP1nipsnap homolog 1 (C. elegans) (ENSG00000184117), score: -0.45 NKIRAS2NFKB inhibitor interacting Ras-like 2 (ENSG00000168256), score: 0.41 NR4A3nuclear receptor subfamily 4, group A, member 3 (ENSG00000119508), score: 0.46 NRAPnebulin-related anchoring protein (ENSG00000197893), score: 0.44 NRF1nuclear respiratory factor 1 (ENSG00000106459), score: 0.43 NSDHLNAD(P) dependent steroid dehydrogenase-like (ENSG00000147383), score: -0.52 NT5C1A5'-nucleotidase, cytosolic IA (ENSG00000116981), score: 0.46 NT5C35'-nucleotidase, cytosolic III (ENSG00000122643), score: 0.57 OGDHLoxoglutarate dehydrogenase-like (ENSG00000197444), score: -0.48 OLFML2Aolfactomedin-like 2A (ENSG00000185585), score: 0.42 ORC2Lorigin recognition complex, subunit 2-like (yeast) (ENSG00000115942), score: -0.46 OTUD1OTU domain containing 1 (ENSG00000165312), score: 0.59 PARP1poly (ADP-ribose) polymerase 1 (ENSG00000143799), score: -0.48 PARS2prolyl-tRNA synthetase 2, mitochondrial (putative) (ENSG00000162396), score: 0.42 PDE3Aphosphodiesterase 3A, cGMP-inhibited (ENSG00000172572), score: 0.46 PDE6Gphosphodiesterase 6G, cGMP-specific, rod, gamma (ENSG00000185527), score: 0.54 PDHBpyruvate dehydrogenase (lipoamide) beta (ENSG00000168291), score: 0.54 PDLIM5PDZ and LIM domain 5 (ENSG00000163110), score: 0.54 PFKFB36-phosphofructo-2-kinase/fructose-2,6-biphosphatase 3 (ENSG00000170525), score: -0.53 PHF5APHD finger protein 5A (ENSG00000100410), score: 0.45 PLEKHA7pleckstrin homology domain containing, family A member 7 (ENSG00000166689), score: 0.49 PLNphospholamban (ENSG00000198523), score: 0.53 POLR3Apolymerase (RNA) III (DNA directed) polypeptide A, 155kDa (ENSG00000148606), score: -0.54 POLR3Kpolymerase (RNA) III (DNA directed) polypeptide K, 12.3 kDa (ENSG00000161980), score: 0.49 PPARGC1Bperoxisome proliferator-activated receptor gamma, coactivator 1 beta (ENSG00000155846), score: 0.41 PPFIBP2PTPRF interacting protein, binding protein 2 (liprin beta 2) (ENSG00000166387), score: 0.51 PPIP5K2diphosphoinositol pentakisphosphate kinase 2 (ENSG00000145725), score: 0.63 PPP1R13Bprotein phosphatase 1, regulatory (inhibitor) subunit 13B (ENSG00000088808), score: 0.44 PPTC7PTC7 protein phosphatase homolog (S. cerevisiae) (ENSG00000196850), score: 0.64 PRKAB2protein kinase, AMP-activated, beta 2 non-catalytic subunit (ENSG00000131791), score: 0.43 PRKAG2protein kinase, AMP-activated, gamma 2 non-catalytic subunit (ENSG00000106617), score: 0.52 PROSCproline synthetase co-transcribed homolog (bacterial) (ENSG00000147471), score: 0.42 PSEN1presenilin 1 (ENSG00000080815), score: -0.47 PTRFpolymerase I and transcript release factor (ENSG00000177469), score: 0.5 RAB10RAB10, member RAS oncogene family (ENSG00000084733), score: 0.44 RBM20RNA binding motif protein 20 (ENSG00000203867), score: 0.54 RBM24RNA binding motif protein 24 (ENSG00000112183), score: 0.54 RBPJLrecombination signal binding protein for immunoglobulin kappa J region-like (ENSG00000124232), score: 0.73 RBPMS2RNA binding protein with multiple splicing 2 (ENSG00000166831), score: 0.53 RGS5regulator of G-protein signaling 5 (ENSG00000143248), score: 0.5 RHAGRh-associated glycoprotein (ENSG00000112077), score: 0.47 RILPL1Rab interacting lysosomal protein-like 1 (ENSG00000188026), score: 0.55 RMND5Arequired for meiotic nuclear division 5 homolog A (S. cerevisiae) (ENSG00000153561), score: 0.42 RNF166ring finger protein 166 (ENSG00000158717), score: 0.56 ROR1receptor tyrosine kinase-like orphan receptor 1 (ENSG00000185483), score: 0.53 RORARAR-related orphan receptor A (ENSG00000069667), score: -0.46 RPL3Lribosomal protein L3-like (ENSG00000140986), score: 0.53 RRADRas-related associated with diabetes (ENSG00000166592), score: 0.48 RTN4IP1reticulon 4 interacting protein 1 (ENSG00000130347), score: 0.43 SCARF1scavenger receptor class F, member 1 (ENSG00000074660), score: 0.42 SEPT2septin 2 (ENSG00000168385), score: -0.47 SGCGsarcoglycan, gamma (35kDa dystrophin-associated glycoprotein) (ENSG00000102683), score: 0.43 SH3BP4SH3-domain binding protein 4 (ENSG00000130147), score: 0.42 SIRT1sirtuin 1 (ENSG00000096717), score: 0.45 SLBPstem-loop binding protein (ENSG00000163950), score: -0.48 SLC16A7solute carrier family 16, member 7 (monocarboxylic acid transporter 2) (ENSG00000118596), score: 0.48 SLC25A28solute carrier family 25, member 28 (ENSG00000155287), score: 0.45 SLC25A3solute carrier family 25 (mitochondrial carrier; phosphate carrier), member 3 (ENSG00000075415), score: 0.48 SLC8A1solute carrier family 8 (sodium/calcium exchanger), member 1 (ENSG00000183023), score: 0.53 SLC9A3R1solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 1 (ENSG00000109062), score: -0.52 SMPXsmall muscle protein, X-linked (ENSG00000091482), score: 0.49 SMYD1SET and MYND domain containing 1 (ENSG00000115593), score: 0.58 SNX13sorting nexin 13 (ENSG00000071189), score: 0.48 SORBS1sorbin and SH3 domain containing 1 (ENSG00000095637), score: 0.42 SOS2son of sevenless homolog 2 (Drosophila) (ENSG00000100485), score: 0.41 SQLEsqualene epoxidase (ENSG00000104549), score: -0.54 SS18L1synovial sarcoma translocation gene on chromosome 18-like 1 (ENSG00000184402), score: -0.56 ST8SIA2ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 2 (ENSG00000140557), score: 0.7 STRN3striatin, calmodulin binding protein 3 (ENSG00000196792), score: 0.43 SYDE2synapse defective 1, Rho GTPase, homolog 2 (C. elegans) (ENSG00000097096), score: 0.62 TANC1tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 (ENSG00000115183), score: 0.49 TBX20T-box 20 (ENSG00000164532), score: 0.63 TBX5T-box 5 (ENSG00000089225), score: 0.57 TCF12transcription factor 12 (ENSG00000140262), score: 0.56 TCF3transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) (ENSG00000071564), score: 0.49 TECRLtrans-2,3-enoyl-CoA reductase-like (ENSG00000205678), score: 0.66 TECTBtectorin beta (ENSG00000119913), score: 0.43 TIE1tyrosine kinase with immunoglobulin-like and EGF-like domains 1 (ENSG00000066056), score: 0.47 TMEM18transmembrane protein 18 (ENSG00000151353), score: -0.45 TMEM194Atransmembrane protein 194A (ENSG00000166881), score: 0.57 TMEM26transmembrane protein 26 (ENSG00000196932), score: 0.49 TMEM9transmembrane protein 9 (ENSG00000116857), score: -0.49 TMOD1tropomodulin 1 (ENSG00000136842), score: 0.46 TNNC1troponin C type 1 (slow) (ENSG00000114854), score: 0.48 TNNT2troponin T type 2 (cardiac) (ENSG00000118194), score: 0.63 TPH2tryptophan hydroxylase 2 (ENSG00000139287), score: 0.48 TRAF3TNF receptor-associated factor 3 (ENSG00000131323), score: -0.49 TRIM55tripartite motif-containing 55 (ENSG00000147573), score: 0.64 TRIM63tripartite motif-containing 63 (ENSG00000158022), score: 0.59 TSC22D2TSC22 domain family, member 2 (ENSG00000196428), score: 0.42 TXLNBtaxilin beta (ENSG00000164440), score: 0.54 UBXN2AUBX domain protein 2A (ENSG00000173960), score: 0.54 UNC119unc-119 homolog (C. elegans) (ENSG00000109103), score: -0.58 UNC50unc-50 homolog (C. elegans) (ENSG00000115446), score: -0.53 UQCR11ubiquinol-cytochrome c reductase, complex III subunit XI (ENSG00000127540), score: 0.48 UQCRC1ubiquinol-cytochrome c reductase core protein I (ENSG00000010256), score: 0.51 UQCRC2ubiquinol-cytochrome c reductase core protein II (ENSG00000140740), score: 0.54 USP28ubiquitin specific peptidase 28 (ENSG00000048028), score: 0.58 VPS33Bvacuolar protein sorting 33 homolog B (yeast) (ENSG00000184056), score: 0.49 WFDC1WAP four-disulfide core domain 1 (ENSG00000103175), score: 0.46 XPNPEP3X-prolyl aminopeptidase (aminopeptidase P) 3, putative (ENSG00000196236), score: 0.46 ZNF366zinc finger protein 366 (ENSG00000178175), score: 0.45
| Id | species | tissue | sex | individual |
|---|---|---|---|---|
| gga_ht_f_ca1 | gga | ht | f | _ |
| mdo_ht_m_ca1 | mdo | ht | m | _ |
| gga_ht_m_ca1 | gga | ht | m | _ |
| mdo_ht_f_ca1 | mdo | ht | f | _ |